|
|
 |
| јвторизаци€ |
|
|
 |
| ѕоиск по указател€м |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Branden C., Tooze J. Ч Introduction to protein structure |
|
|
 |
| ѕредметный указатель |
Sliding filament model 291 291F
Smith, George 361
Snake venom 26 26F
Solar cells 240 244
Somatic hypermutation 303
Specificity constant 206
Specificity pocket 209
Specificity pocket, chymotrypsin 211 211F
Specificity pocket, engineered mutations 213Ч215
Specificity pocket, hydrophobic, in subtilisin 217
Specificity pocket, preferential cleavage conferred by side chains 212Ч213
Spectrin 36
Spider dragline fibers 289
Spider silk 289Ч290 290F
SpidersТ webs 283
Spin, of nuclei 387
Spiral see УHelical wheelФ
Spongiform encephalopathies 113 288
Sprang, Stephen 256 262
Src tyrosine kinase 275Ч277
Src tyrosine kinase, active and inactive forms 278
Src tyrosine kinase, helix 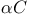 278 278
Src tyrosine kinase, phosphorylation 275 276
Src tyrosine kinase, regulation 278F
Src tyrosine kinase, SH2 domain 273F 275Ч277
Src tyrosine kinase, SH2 domain, linker region with N-terminal domain 277
Src tyrosine kinase, SH3 domain 275Ч277
Src tyrosine kinase, structure 276F 277F
Src-homology-2 see УSH2 domainФ
Src-homology-3 see УSH3 domainФ
Stability in protein engineering see УProtein engineeringФ
Staggered conformations 10Ч11 10F
Staphylococcus nuclease 27 27F
Steitz, Thomas 58F 146 245
Stemmer, Willem 365
Stereochemical data, prediction of secondary structure 351
Strandberg, Bror 329
Streptomyces lividans 232
Structural hierarchy, of proteins 28Ч29
Stuart, David 333
Substrate specificity, chymotrypsin family 212Ч213
Substrate specificity, trypsin mutation 213 215
Substrate-assisted catalysis 218Ч219 218F
Substrates, transition state 207 207F
Subtilisin 28F 215
Subtilisin,  motif and handedness 217 motif and handedness 217
Subtilisin, active sites 216Ч217 216F
Subtilisin, amino acids 215
Subtilisin, catalysis without catalytic triad 217Ч218
Subtilisin, catalytic triad 216 217
Subtilisin, evolution 210
Subtilisin, protein engineering 215 217 219
Subtilisin, structural anomaly and functional effects 217
Subtilisin, structure 215 216F
Subtilisin, substrate-assisted catalysis 218Ч219
Subtilisin, transition-state stabilization 217
Sugar-binding proteins 62F 6363F
Superbarrel, neuraminidase 72
Superfamily, chymotrypsin 210 212
Superfamily, retinol-binding protein (RBP) 70
Superoxide dismutase (SOD) 67F
Supersaturated solutions 375
Supersecondary structures see УMotifsФ
SV40 326 341Ч343
SV40, structure 342 342F
Swinging cross-bridge model 292 292F 296F
Swinging cross-bridge model, myosin structure supporting 295Ч296
Switch points, topological 59 60
Switch regions 256
Switch regions,  activation 257Ч259 activation 257Ч259
Switch regions, myosin S1 fragment 294
Synchrotrons 376 377 384
T (tense) state 113Ч114
T-cell receptors (TCR) 299 300 316Ч317
T-cell receptors (TCR), antigen-binding site 317F
T-cell receptors (TCR), gene rearrangements 316F 317
T-cell receptors (TCR), hypervariable regions 316 317 318F
T-cell receptors (TCR), MHC-peptide complexes as ligands 318 318F
T-cell receptors (TCR), variable and constant regions 316Ч317
T-cells, activation 312Ч313
T-loop 108 109F
T4 bacteriophage see УBacteriophage T4Ф
Tandem repeats, leucine-rich in in  -horseshoe folds 56 -horseshoe folds 56
Tandem repeats, retinoid X receptor recognizing 185Ч186
TATA box 151 151F 155
TATA box, consensus sequence 154F
TATA box, DNA deformation 155Ч157
TATA box, TBP binding, hydrogen bonds 157 158
TATA box, TBP binding, hydrophobic interactions 157Ч158
TATA box, TBP binding, sequence-specific 157Ч158 157F
TATA box, TBP complexes with 154
TATA box, TBP preference for 157 158
TATA box-binding protein (TBP) 151 152 152F 153Ч154
TATA box-binding protein (TBP), amino acid sequences 153
TATA box-binding protein (TBP), binding to minor groove of DNA 155Ч157 156F
TATA box-binding protein (TBP), C-terminal domain 153Ч154
TATA box-binding protein (TBP), DNA deformation after binding 155Ч157 156F
TATA box-binding protein (TBP), DNA-binding sites as  sheet 154 sheet 154
TATA box-binding protein (TBP), isolation 153
TATA box-binding protein (TBP), motifs 158
TATA box-binding protein (TBP), mutants 153Ч154
TATA box-binding protein (TBP), N-terminal segment 154
TATA box-binding protein (TBP), preference for TATA box 157 158
TATA box-binding protein (TBP), promoter complex 159
TATA box-binding protein (TBP), side chains 154 157
TATA box-binding protein (TBP), TATA box complexes 154
TATA box-binding protein (TBP), TFIIA and TFIIB binding 159
TATA box-binding protein (TBP), ubiquity 153Ч154
Taylor, Susan 278
TBP see УTATA box-binding protein (TBP)Ф
TBP-associated factors (TAFs) 152 153
Temperature factor (B) 383
Temperature, melting (Tm) 354 356F
Tertiary structure of proteins 3F 28 29
Tertiary structure of proteins, local secondary structure imposed by 351
Tertiary structure of proteins, molten globular state 89
Tertiary structure of proteins, prediction, secondary structure knowledge required 350Ч351
Tertiary structure of proteins, protein engineering 347Ч348 (see also УDomainsФ УMotifsФ УThree-dimensional
TFIIA see УTranscription factor TFIIAФ
Thioredoxin, NMR and x-rav crystallography comparison 391
Thioredoxin, phosducin homology 265Ч266
Thioredoxin, structure 20F 97
Threading methods 353Ч354
Three-dimensional model, homologous proteins 348
Three-dimensional structure of proteins 3 4
Three-dimensional structure of proteins, disulfide bridge stabilization of 8
Three-dimensional structure of proteins, prediction 89 349 349F
Three-dimensional structure of proteins, similar with different amino acid sequences 352 (see also УDomainsФ УMotifsФ УTertiary
three-dimensional structures 303Ч304 304F
Threonine, Ras and  proteins 256 proteins 256
Threonine, structure 6F
Thymine, in DNA, hydrogen bonds 123F
Thyroid hormone receptor 185 186F
TIM barrel 47 267
Tissue factor-factor Vila (TF-FVIIa) 361 362 362T
Titin 290Ч291
Tm (melting temperature) 354 356F
Tobacco mosaic virus 326F
Tobacco mosaic virus, coat protein 37
Tomato bushy stunt virus 331Ч332 332F
Tomato bushy stunt virus, capsid structure 331Ч332 332F
Tomato bushy stunt virus, jelly roll structure 335F
Tomato bushy stunt virus, S and  domains 332 332F domains 332 332F
Tomato bushy stunt virus, size 332
Tomato bushy stunt virus, subnits recognizing RNA 332Ч333 333F
Topological switch points 59 60
Topology diagram 23
Topology diagram,  domains 48F domains 48F
Topology diagram,  motif 28F motif 28F
Topology diagram,  -crystallin 74F 75F -crystallin 74F 75F
| Topology diagram, carboxypeptidase 61F
Topology diagram, chymotrypsin 211F
Topology diagram, complex motifs 31 31F
Topology diagram, fibronectin type III domain 319F
Topology diagram, Greek key motifs 27F
Topology diagram, immunoglobulin 304F
Topology diagram, jelly roll motif 78F
Topology diagram, neuraminidase 71F
Topology diagram, open twisted  structures 58F structures 58F
Topology diagram, pS3 DNA-binding domain 168
Topology diagram, Ras proteins 254F
Topology diagram, subtilisin 217F
Topology diagram, TATA box-binding protein 155F
Topology diagram, up-and-down  barrels 68F barrels 68F
Topology diagram, uses and advantages 76
Trans-peptide 98 98F
trans-retinoic acid (RAR) receptor 185 186F
Transcription factor 151Ч174 175Ч203
Transcription factor GCN4 see УGCN4Ф
Transcription factor Oct-1 164F 165
Transcription factor PPR1 190Ч191 190F
Transcription factor TFIIA, binding to TBP and DNA 159
Transcription factor TFIIA, domains 159
Transcription factor TFIIB, binding to TBP and DNA 159
Transcription factor TFIIB, domains 159
Transcription factor TFIID 151 152 158
Transcription factor TFIIIA, zinc finger motifs 176 177
Transcription factor, classes 152
Transcription factor, definition 151
Transcription factor, domains 153
Transcription factor, eucaryotic 175 191
Transcription factor, evolution 202
Transcription factor, families 153 175Ч203
Transcription factor, functions of polypeptide chains 152Ч153
Transcription factor, genes encoding, cloning 153
Transcription factor, helix-loop-helix (HLH) family 39
Transcription factor, heterodimer binding to DNA 163 163F
Transcription factor, homeodomains see УHomeodomainsФ
Transcription factor, immunoglobulin fold in 168Ч169
Transcription factor, leucine zipper motifs 191Ч193
Transcription factor, POU region 164F
Transcription factor, procaryotic 175
Transcription factor, selectivity, homeodomains 162Ч164
Transcription factor, zinc-containing motifs see УZinc-containing motifsФ
Transcription, activation by CAP-induced DNA bending 146Ч147
Transcription, activation, model 152F
Transcription, activation, protein-protein interactions 152Ч153
Transcription, DNA elements involved 151F
Transcription, initiation complex 153
Transcription, initiation, TBP binding to TATA box and 158
Transcription, preinitiation complex 151 152 159
Transcription, regulation 152
Transcription, regulation, eucaryotes 151
Transducin 256
Transducin,  structure 256 256F structure 256 256F
Transducin, 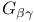 structure 262 262F 263 263F structure 262 262F 263 263F
Transducin, activation by rhodopsin 265
Transition states 206
Transmembrane  helices 226Ч227 helices 226Ч227
Transmembrane  helices, bacteriorhodopsin 226Ч227 226F helices, bacteriorhodopsin 226Ч227 226F
Transmembrane  helices, hydropathy plots 245 246F helices, hydropathy plots 245 246F
Transmembrane  helices, LH2 light-harvesting complex 241 241F 242F helices, LH2 light-harvesting complex 241 241F 242F
Transmembrane  helices, no specific interaction with membrane lipids 246Ч247 helices, no specific interaction with membrane lipids 246Ч247
Transmembrane  helices, photosynthetic reaction center 244Ч245 helices, photosynthetic reaction center 244Ч245
Transmembrane  helices, prediction from amino acid sequences 244Ч245 helices, prediction from amino acid sequences 244Ч245
Transmembrane  helices, subunits of photosynthetic reaction center 236Ч237 237F helices, subunits of photosynthetic reaction center 236Ч237 237F
Transmembrane enzyme-linked receptors 271
Transmembrane proteins,  helices 18 helices 18
Transthyretin 288
Transthyretin, twisted  helix 288 288F 289F helix 288 288F 289F
Triangulation numbers (T) 330
Triosephosphate isomerase,  motif 28 28F motif 28 28F
Triosephosphate isomerase,  motifs 30 30F motifs 30 30F
Triosephosphate isomerase, amino acids of  strands 48 50T strands 48 50T
Triosephosphate isomerase, structure 23F
Triosephosphate isomerase, TIM barrel structure 47
Troponin-C 24 26
Troponin-C, helical wheel 17T
trp repressor, conformational change effect 142Ч143
trp repressor, dimerization 142 142F
trp repressor, helix-turn-helix motif 142 142F
trp repressor, structure 142
trp repressor, tryptophan binding 142Ч143 143F
Trypsin, Asp 189-Lys mutation 215
Trypsin, kinetic data 214F
Trypsin, mutant, specificity pocket 214F
Trypsin, preferential cleavage sites 212 213 213F
Trypsin, specificity mechanism 209
Trypsin, specificity pocket 214F
Tryptophan, binding to trp repressor 142Ч143 143F
Tryptophan, operon 142
Tryptophan, structure 7F
Tryptophan, synthesis 142
TTK 179 180F
Tu, elongation factor 255
Tubulin 284
Tumor suppressor gene 166 (see also Уp53Ф)
Tumorigenic mutations 166
Tumorigenic mutations, pS3 regions 170Ч171 170F
Turnover number (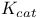 ) 206 ) 206
Two-dimensional crystals, membrane proteins 225Ч226 226F
Tyrosine kinase, receptor 270Ч271 271 272F
Tyrosine kinase, Src see УSrc tyrosine kinaseФ
Tyrosine kinase-associated receptors 270 271 272F
Tyrosine kinase-associated receptors, adaptor modules 272 272F
Tyrosine phosphatases 271
Tyrosine, structure 6F 59
Tyrosyl adenylate 60F
Tyrosyl-tRNA synthetase, domains 59Ч60 59F 60F
Tyrosyl-tRNA synthetase, side chains 61F
Tyrosyl-tRNA synthetase, tyrosine bound to 60 60F
Ubiquinone 236F
Ultraviolet light, Cro formation and lytic cycle 131 133
Unit cell 374
Unwin, Nigel 226
Up-and-down  barrels 68 68Ч71 68F barrels 68 68Ч71 68F
Up-and-down  barrels, porin channels 229Ч230 230F barrels, porin channels 229Ч230 230F
Up-and-down  barrels, SH3 domain 274 275F barrels, SH3 domain 274 275F
Up-and-down  sheets, in neuraminidase 70Ч71 71F sheets, in neuraminidase 70Ч71 71F
Upstream-activating sequence (UAS), GAL4 188
Urokinase 29F
Valegard, Karin 339
Valine, recognition helix of glucocorticoid receptor 184Ч185
Valine, staggered conformations 10Ч11 10F
Valine, structure 6F
Vibrio cholerae 254
Vimentin 287F
Viruses 325Ч345
Viruses, capsid 325 326
Viruses, capsid, polypeptide chains 325
Viruses, enveloped 325
Viruses, genomes 325 326 330
Viruses, infections 325
Viruses, jelly roll structures 335Ч337 335F 336F
Viruses, pentamers of coat protein 341Ч343 342F
Viruses, simplest 328Ч239
Viruses, sizes and shapes 326F
Viruses, spherical 325Ч345
Viruses, spherical, complex 329Ч331
Viruses, spherical, icosahedral symmetry 327 327F
Viruses, T=3 structure 330 330F
Viruses, T=3 structure, plant viruses 331Ч332 337
Viruses, T=4 structure 331 331F
Viruses, T=4 structure, Sindbis virus 341
Viruses, T=7 design 341Ч342 342F
Vitamin A (retinol) 68 68F 69F
Vitamin D receptor 185 186F
Vitamin D-resistant rickets 184
Vitronectin 113
Water in  activation 258 activation 258
|
|
 |
| –еклама |
 |
|
|
 |
|
ќ проекте
|
|
ќ проекте



 motif and handedness
motif and handedness  activation
activation  -horseshoe folds
-horseshoe folds  sheet
sheet  -crystallin
-crystallin 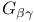 structure
structure  helices
helices  motifs
motifs 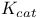 )
)