|
|
 |
| јвторизаци€ |
|
|
 |
| ѕоиск по указател€м |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Branden C., Tooze J. Ч Introduction to protein structure |
|
|
 |
| ѕредметный указатель |
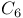 -zinc cluster family 190Ч191 190F 202 -zinc cluster family 190Ч191 190F 202
 fragment 224Ч225 fragment 224Ч225
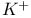 leak channels see УPotassium ( leak channels see УPotassium (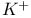 ) channelsФ ) channelsФ
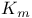 values 206 213 214 values 206 213 214
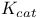 values 206 213 214 values 206 213 214
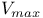 206 206
 domains 31 35Ч46 domains 31 35Ч46
 domains, coiled-coil domains, coiled-coil  helices 35Ч37 helices 35Ч37
 domains, doughnut-shaped 39Ч40 39F domains, doughnut-shaped 39Ч40 39F
 domains, evolutionary conservation 41Ч42 domains, evolutionary conservation 41Ч42
 domains, four-helix bundle 37Ч39 38F domains, four-helix bundle 37Ч39 38F
 domains, globin fold 22F 35 40 40F domains, globin fold 22F 35 40 40F
 domains, helix movements and side-chain mutations 43 domains, helix movements and side-chain mutations 43
 domains, hydrophobic interior 35 42Ч43 domains, hydrophobic interior 35 42Ч43
 domains, packing of domains, packing of  helices 40Ч41 41F 42F helices 40Ч41 41F 42F
 domains, size and complexity 39Ч40 domains, size and complexity 39Ч40
 domains, tyrosyl-tRNA synthetase 59Ч60 59F 60F domains, tyrosyl-tRNA synthetase 59Ч60 59F 60F
 helix (helices) 14Ч19 15F helix (helices) 14Ч19 15F
 helix (helices) in helix (helices) in  domains 31 35 domains 31 35
 helix (helices) in helix (helices) in  structures, barrels 49 49F structures, barrels 49 49F
 helix (helices) in helix (helices) in  structures, horseshoe folds 55 56F structures, horseshoe folds 55 56F
 helix (helices) in helix (helices) in  structures, twisted open sheets 56Ч57 57F structures, twisted open sheets 56Ч57 57F
 helix (helices) in helix (helices) in  motif 27Ч28 28F motif 27Ч28 28F
 helix (helices) in helix-turn-helix motif 129 helix (helices) in helix-turn-helix motif 129
 helix (helices) in membrane-bound proteins 35 helix (helices) in membrane-bound proteins 35
 helix (helices) in zinc motif of glucocorticoid receptor 184Ч185 helix (helices) in zinc motif of glucocorticoid receptor 184Ч185
 helix (helices), helix (helices), 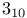 helix 15 177 helix 15 177
 helix (helices), helix (helices), 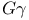 263 263
 helix (helices), 6-F helix 116 helix (helices), 6-F helix 116
 helix (helices), amino acids preferred 16Ч18 helix (helices), amino acids preferred 16Ч18
 helix (helices), b/HLH/zip family 200 helix (helices), b/HLH/zip family 200
 helix (helices), bacteriophage MS2 subunits 339 helix (helices), bacteriophage MS2 subunits 339
 helix (helices), barnase folding intermediate 94 94F helix (helices), barnase folding intermediate 94 94F
 helix (helices), buried and exposed types and sequences 17T helix (helices), buried and exposed types and sequences 17T
 helix (helices), C- and N-termtni 16 helix (helices), C- and N-termtni 16
 helix (helices), calmodulin 110 helix (helices), calmodulin 110
 helix (helices), CDK2 (PSTAIRE helix) 107F 108 108F 109F helix (helices), CDK2 (PSTAIRE helix) 107F 108 108F 109F
 helix (helices), coiled-coll 35Ч37 35F 36F helix (helices), coiled-coll 35Ч37 35F 36F
 helix (helices), coiled-coll, GAL4 187 helix (helices), coiled-coll, GAL4 187
 helix (helices), coiled-coll, heptad repeats 35Ч37 36F 192 286 helix (helices), coiled-coll, heptad repeats 35Ч37 36F 192 286
 helix (helices), coiled-coll, leucine zipper dimerization 192 helix (helices), coiled-coll, leucine zipper dimerization 192
 helix (helices), coiled-coll, oligomers of fibrous proteins 286Ч287 helix (helices), coiled-coll, oligomers of fibrous proteins 286Ч287
 helix (helices), coiled-coll, stabilization by salt bridges 36 37F helix (helices), coiled-coll, stabilization by salt bridges 36 37F
 helix (helices), conversion from p sheet (protein design) 368Ч370 369F 369T helix (helices), conversion from p sheet (protein design) 368Ч370 369F 369T
 helix (helices), Cro protein 132 helix (helices), Cro protein 132
 helix (helices), dipole moment 16 helix (helices), dipole moment 16
 helix (helices), dipole moment, stabilizing to increase protein stability 357Ч358 357F helix (helices), dipole moment, stabilizing to increase protein stability 357Ч358 357F
 helix (helices), DsbA 97 helix (helices), DsbA 97
 helix (helices), electron-density maps 381F 382 helix (helices), electron-density maps 381F 382
 helix (helices), F-actin 293 helix (helices), F-actin 293
 helix (helices), four-helix bundle 37Ч39 38F helix (helices), four-helix bundle 37Ч39 38F
 helix (helices), GAL4 187 helix (helices), GAL4 187
 helix (helices), globin fold 40 40F helix (helices), globin fold 40 40F
 helix (helices), globular proteins 15 helix (helices), globular proteins 15
 helix (helices), GroEL domain 100 102 helix (helices), GroEL domain 100 102
 helix (helices), growth hormone 267 267F helix (helices), growth hormone 267 267F
 helix (helices), handedness 16 285 helix (helices), handedness 16 285
 helix (helices), helical wheel 17T helix (helices), helical wheel 17T
 helix (helices), hemagglutinin 79 helix (helices), hemagglutinin 79
 helix (helices), homeodomains 160 helix (helices), homeodomains 160
 helix (helices), hydrogen bonds 15 helix (helices), hydrogen bonds 15
 helix (helices), hydrophilic and hydrophobic residues 17 helix (helices), hydrophilic and hydrophobic residues 17
 helix (helices), lambda repressor 133 helix (helices), lambda repressor 133
 helix (helices), length 15 helix (helices), length 15
 helix (helices), leucine zipper motif 192 192F helix (helices), leucine zipper motif 192 192F
 helix (helices), ligand-binding 16 helix (helices), ligand-binding 16
 helix (helices), location in proteins 17 helix (helices), location in proteins 17
 helix (helices), loop region connections 21 helix (helices), loop region connections 21
 helix (helices), membrane proteins 223 helix (helices), membrane proteins 223
 helix (helices), motifs using 24 helix (helices), motifs using 24
 helix (helices), movements and side-chain mutations 43 helix (helices), movements and side-chain mutations 43
 helix (helices), MyoD 197 198Ч199 helix (helices), MyoD 197 198Ч199
 helix (helices), myosin 294 helix (helices), myosin 294
 helix (helices), p53 169Ч170 helix (helices), p53 169Ч170
 helix (helices), p53, domain 167 helix (helices), p53, domain 167
 helix (helices), packing arrangements 40Ч41 helix (helices), packing arrangements 40Ч41
 helix (helices), packing in helix (helices), packing in  domains 40Ч41 41F 42F domains 40Ч41 41F 42F
 helix (helices), prediction of secondary structure 352 helix (helices), prediction of secondary structure 352
 helix (helices), proline in 16Ч17 helix (helices), proline in 16Ч17
 helix (helices), Ramachandran plot 10 helix (helices), Ramachandran plot 10
 helix (helices), Ras protein 255 helix (helices), Ras protein 255
 helix (helices), recognition helix (helices), recognition  helix 134 helix 134
 helix (helices), residues per turn 15 helix (helices), residues per turn 15
 helix (helices), ridges and grooves geometry 40Ч41 41F helix (helices), ridges and grooves geometry 40Ч41 41F
 helix (helices), Rop protein 38Ч39 39F helix (helices), Rop protein 38Ч39 39F
 helix (helices), schematic diagram representation 23 helix (helices), schematic diagram representation 23
 helix (helices), secondary structure of proteins 14Ч16 helix (helices), secondary structure of proteins 14Ч16
 helix (helices), serpins 111Ч112 helix (helices), serpins 111Ч112
 helix (helices), SH2 domain 273 helix (helices), SH2 domain 273
 helix (helices), side chains 17 43 helix (helices), side chains 17 43
 helix (helices), subtilisin 215 helix (helices), subtilisin 215
 helix (helices), SV40 343 343F helix (helices), SV40 343 343F
 helix (helices), TATA box-binding protein 154 helix (helices), TATA box-binding protein 154
 helix (helices), transducin helix (helices), transducin  256 257 256 257
 helix (helices), transmembrane see УTransmembrane helix (helices), transmembrane see УTransmembrane  helicesФ helicesФ
 helix (helices), transmembrane proteins 18 223 helix (helices), transmembrane proteins 18 223
 helix (helices), trp repressor 142 helix (helices), trp repressor 142
 helix (helices), variations 15 helix (helices), variations 15
 helix (helices), x-ray diffraction data 382 helix (helices), x-ray diffraction data 382
 -Lytic protease 92 -Lytic protease 92
 -Lytic protease, folding with prosegment 92 -Lytic protease, folding with prosegment 92
 barrels 44 48Ч49 barrels 44 48Ч49
 barrels in pyruvate kinase 51Ч52 51F barrels in pyruvate kinase 51Ч52 51F
 barrels, active sites 53Ч54 53F barrels, active sites 53Ч54 53F
 barrels, active sites, prediction 57 59 barrels, active sites, prediction 57 59
 barrels, amino acid residues 48 50T barrels, amino acid residues 48 50T
 barrels, branched hydrophobic side chains 49Ч51 barrels, branched hydrophobic side chains 49Ч51
 barrels, double 52Ч53 52F barrels, double 52Ч53 52F
 barrels, enzymes containing 48Ч49 barrels, enzymes containing 48Ч49
 barrels, evolution of new enzyme activities 54Ч55 barrels, evolution of new enzyme activities 54Ч55
 barrels, methylmalonyl-coenzyme A mutase 50Ч51 50T barrels, methylmalonyl-coenzyme A mutase 50Ч51 50T
 barrels, parallel barrels, parallel  strands 48 49F 306 strands 48 49F 306
 domains 32 47Ч65 48F domains 32 47Ч65 48F
 domains, active site prediction 57 59 domains, active site prediction 57 59
 domains, arabinose-binding protein 62Ч63 62F domains, arabinose-binding protein 62Ч63 62F
 horseshoe folds 55 56F horseshoe folds 55 56F
 proteins/structures, proteins/structures,  helix relationship 84 helix relationship 84
 proteins/structures, carboxypeptidase 60Ч62 61F proteins/structures, carboxypeptidase 60Ч62 61F
 proteins/structures, classes 47Ч48 proteins/structures, classes 47Ч48
 proteins/structures, phosphofructokinase 115 115F proteins/structures, phosphofructokinase 115 115F
 proteins/structures, Ras protein 255 proteins/structures, Ras protein 255
 proteins/structures, subtilisin 215 (see also У proteins/structures, subtilisin 215 (see also У domainsФ) domainsФ)
 sheets, open twisted 47 56Ч57 sheets, open twisted 47 56Ч57
 sheets, open twisted, active-site crevice 56Ч57 57F 63 sheets, open twisted, active-site crevice 56Ч57 57F 63
 sheets, open twisted, two domains in proteins 63 sheets, open twisted, two domains in proteins 63
 sheets, types 56Ч57 sheets, types 56Ч57
 twisted open-sheet structures 47 56Ч57 twisted open-sheet structures 47 56Ч57
 twisted open-sheet structures, twisted open-sheet structures,  domain with in tyrosyl-tRNA synthetase 59Ч60 59F 60F domain with in tyrosyl-tRNA synthetase 59Ч60 59F 60F
 twisted open-sheet structures, carboxypeptidase 60Ч62 61F twisted open-sheet structures, carboxypeptidase 60Ч62 61F
 twisted open-sheet structures, classes 47 twisted open-sheet structures, classes 47
 twisted open-sheet structures, G-actin 293 293F twisted open-sheet structures, G-actin 293 293F
 twisted open-sheet structures, mixed twisted open-sheet structures, mixed  sheet with 60Ч62 61F sheet with 60Ч62 61F
 twisted open-sheet structures, pyruvate kinase 51Ч52 51F twisted open-sheet structures, pyruvate kinase 51Ч52 51F
 twisted open-sheet structures, two similar in arabinose-binding protein 62Ч63 62F twisted open-sheet structures, two similar in arabinose-binding protein 62Ч63 62F
 twisted open-sheet structures, tyrosyl-tRNA synthetase 59Ч60 59F 60F У twisted open-sheet structures, tyrosyl-tRNA synthetase 59Ч60 59F 60F У
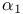 -Antitrypsin 110Ч111 -Antitrypsin 110Ч111
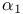 -Antitrypsin, active, cleaved and latent forms 112 112F -Antitrypsin, active, cleaved and latent forms 112 112F
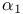 -Antitrypsin, deficiency 113 -Antitrypsin, deficiency 113
 Amyloid 290 Amyloid 290
 barrels, antiparallel see УAntiparallel barrels, antiparallel see УAntiparallel  barrelsФ barrelsФ
 domains 31 domains 31
 sheets 14 19Ч20 47 48 sheets 14 19Ч20 47 48
 sheets in sheets in  twisted open sheets 56Ч57 57F twisted open sheets 56Ч57 57F
 sheets in sheets in  helix 84Ч85 84F 85F helix 84Ч85 84F 85F
 sheets in antiparallel sheets in antiparallel  structures 67 67F structures 67 67F
 sheets, active sites in sheets, active sites in  structures 57 59 structures 57 59
 sheets, antiparallel see УAntiparallel sheets, antiparallel see УAntiparallel  sheetsФ sheetsФ
 sheets, barnase folding intermediate 94 94F sheets, barnase folding intermediate 94 94F
 sheets, DNA-binding site of TATA box-binding protein 154 sheets, DNA-binding site of TATA box-binding protein 154
 sheets, four-stranded 30Ч31 31F 68 sheets, four-stranded 30Ч31 31F 68
|  sheets, GroEL domain 100 sheets, GroEL domain 100
 sheets, hairpin sheets, hairpin  motifs in 26 26F motifs in 26 26F
 sheets, immunoglobulins 307 sheets, immunoglobulins 307
 sheets, jelly roll barrel 78 sheets, jelly roll barrel 78
 sheets, MHC class I molecule 314 sheets, MHC class I molecule 314
 sheets, mixed 20 20F sheets, mixed 20 20F
 sheets, mixed, carboxypeptidase 60Ч62 61F sheets, mixed, carboxypeptidase 60Ч62 61F
 sheets, mixed, thioredoxin 97 sheets, mixed, thioredoxin 97
 sheets, open structures, topologies 57 58F sheets, open structures, topologies 57 58F
 sheets, open twisted 47 48 48F sheets, open twisted 47 48 48F
 sheets, parallel 19 19F sheets, parallel 19 19F
 sheets, parallel, sheets, parallel,  -horseshoe folds 55 56F -horseshoe folds 55 56F
 sheets, parallel, subtilisin 215 216F sheets, parallel, subtilisin 215 216F
 sheets, parallel, topology diagram 24F (see also У sheets, parallel, topology diagram 24F (see also У strandsФ) strandsФ)
 sheets, pleated 19 19F sheets, pleated 19 19F
 sheets, silk fibroins 289 290F sheets, silk fibroins 289 290F
 sheets, topology diagrams 23 24F sheets, topology diagrams 23 24F
 sheets, twisted 20 20F sheets, twisted 20 20F
 sheets, twisted, transthyretin 288 288F 289F sheets, twisted, transthyretin 288 288F 289F
 sheets, up-and-down see УUp-and-down sheets, up-and-down see УUp-and-down  sheetsФ sheetsФ
 sheets, zinc finger motif binding 178 sheets, zinc finger motif binding 178
 strand insertion in latent serpin 112 112F strand insertion in latent serpin 112 112F
 strand-loop-helix 184 strand-loop-helix 184
 strand-turn- strand-turn- -helix motif, p53 domain 167 -helix motif, p53 domain 167
 strands 19 strands 19
 strands in strands in  helix 84 helix 84
 strands in hairpin strands in hairpin  motif 26Ч27 26F motif 26Ч27 26F
 strands, antiparallel see УAntiparallel strands, antiparallel see УAntiparallel  strandsФ УAntiparallel strandsФ УAntiparallel
 strands, barrel or sheet structures 47Ч48 strands, barrel or sheet structures 47Ч48
 strands, change of active to latent form of serpins 112 112F strands, change of active to latent form of serpins 112 112F
 strands, conversion to a structure (protein design) 368Ч370 369F 369T strands, conversion to a structure (protein design) 368Ч370 369F 369T
 strands, Cro protein 132F strands, Cro protein 132F
 strands, hairpin loops 21Ч22 21F strands, hairpin loops 21Ч22 21F
 strands, hydrogen bonds 19 strands, hydrogen bonds 19
 strands, immunoglobulins 304 304F 305 308F strands, immunoglobulins 304 304F 305 308F
 strands, interactions 19 strands, interactions 19
 strands, jelly roll barrel formation 77Ч78 77F strands, jelly roll barrel formation 77Ч78 77F
 strands, loop region connections 21 strands, loop region connections 21
 strands, membrane proteins 223 strands, membrane proteins 223
 strands, p53 167 168 169 strands, p53 167 168 169
 strands, parallel in strands, parallel in  barrels 48 49F 306 barrels 48 49F 306
 strands, parallel, strands, parallel,  motif 27Ч28 28F motif 27Ч28 28F
 strands, parallel, active site of carboxypeptidase 62 strands, parallel, active site of carboxypeptidase 62
 strands, parallel, barrels or sheet structures 47Ч48 strands, parallel, barrels or sheet structures 47Ч48
 strands, parallel, subtilisin 215 (see also У strands, parallel, subtilisin 215 (see also У sheetФ) sheetФ)
 strands, phosphofructokinase 116 strands, phosphofructokinase 116
 strands, pleated 19 strands, pleated 19
 strands, prediction of secondary structure 352 strands, prediction of secondary structure 352
 strands, Ramachandran plot 10 19 strands, Ramachandran plot 10 19
 strands, Ras protein 255 strands, Ras protein 255
 strands, schematic diagram representation 23 strands, schematic diagram representation 23
 strands, superbarrel formation in neuraminidase 72 strands, superbarrel formation in neuraminidase 72
 strands, SV40 343 343F strands, SV40 343 343F
 strands, transmembrane, porins 228Ч229 strands, transmembrane, porins 228Ч229
 strands, transmembrane, prediction 230 strands, transmembrane, prediction 230
 strands, twisted in strands, twisted in  sheet 20 20F sheet 20 20F
 structures 67Ч88 structures 67Ч88
 structures, structures,  -helix 84Ч85 -helix 84Ч85
 structures, antiparallel see УAntiparallel structures, antiparallel see УAntiparallel  structuresФ structuresФ
 -Crystallin, mouse 76 -Crystallin, mouse 76
 -Lactoglobulin 70 -Lactoglobulin 70
 -loop- -loop-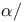 structure 55 56F structure 55 56F
 -loop- -loop- units 31F 68 units 31F 68
 motif 27Ч28 28F motif 27Ч28 28F
 motif in motif in  domains 32 domains 32
 motif in barrel and sheet structures 47 49F motif in barrel and sheet structures 47 49F
 motif, handedness 28 48 49F motif, handedness 28 48 49F
 motif, handedness, left-handed in subtilisin 217 motif, handedness, left-handed in subtilisin 217
 motif, orientation and alignments 47Ч48 49F motif, orientation and alignments 47Ч48 49F
 motif, thioredoxin 97 motif, thioredoxin 97
 motifs 30 30F motifs 30 30F
 unit (hairpin unit (hairpin  motif) 26Ч27 26F motif) 26Ч27 26F
 unit (hairpin unit (hairpin  motif), motif),  structure relationship 84 structure relationship 84
 unit (hairpin unit (hairpin  motif), motif),  strands in 84 strands in 84
 unit (hairpin unit (hairpin  motif), motif),  -helix 84Ч85 -helix 84Ч85
 unit (hairpin unit (hairpin  motif), three-sheet 85 85F 86F motif), three-sheet 85 85F 86F
 unit (hairpin unit (hairpin  motif), twisted, in transthyretin 288 288F 289F motif), twisted, in transthyretin 288 288F 289F
 unit (hairpin unit (hairpin  motif), two-sheet 84 84F motif), two-sheet 84 84F
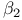 Microglobulin 313 314Ч315 Microglobulin 313 314Ч315
 -Crystallin, coding sequence 76 -Crystallin, coding sequence 76
 -Crystallin, evolution 76 -Crystallin, evolution 76
 -Crystallin, Greek key motifs 74Ч75 75F 76 -Crystallin, Greek key motifs 74Ч75 75F 76
 -Crystallin, two domains 74 74F -Crystallin, two domains 74 74F
 -Crystallin, two domains, amino acid sequences 76 -Crystallin, two domains, amino acid sequences 76
 -Crystallin, two domains, identical topology 75Ч76 -Crystallin, two domains, identical topology 75Ч76
 -Crystallin, two domains, structures 76 -Crystallin, two domains, structures 76
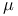 -oxo bridge 11 11F -oxo bridge 11 11F
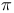 Helix 15 Helix 15
2-Phosphoglycolate 115
3-Methylisoxazole groups 338
3D profile method 353
4-Oxazolinyl phenoxy group 338
6-F Helix 116
A-T base pair 123F
A-T base pair, TATA box 158
Accessory chlorophyll 238 238F 239
Actin 197 290Ч291 291F
Actin, cross-bridges with myosin 291Ч292
Actin, F-actin 293
Actin, fibrous protein 292
Actin, G-actin 293 293F
Actin, myosin complex structure 295 295F
Actin, structure 293Ч295
Activation energy of reactions 206
Activation energy of reactions, enzymes decreasing 206Ч207 207F
Active sites 3F
Active sites, arabinose-binding protein 62F 63
Active sites, carboxypeptidase 62F
Active sites, chymotrypsin 211Ч212 211F 212F
Active sites, crevices 57 59 63
Active sites, neuraminidase 71F 72
Active sites, parallel p strands flanked by antlparallel P strands 62
Active sites, RuBisCo 53F
Active sites, rx/p structures, barrels 53Ч54 53F
Active sites, rx/p structures, domains 57 59
Active sites, rx/p structures, open twisted domains 56 57 57F 59 59F
Active sites, rx/p structures, prediction 57 59
Active sites, serine protease 211Ч212 211F 212F 361
Active sites, subtilisin 216Ч217 216F
Active sites, tyrosyl-tRNA synthetase 59Ч60 59F 60F
Acyl-enzyme intermediates 208 208F
Additivity principle 362
Adenine in DNA, hydrogen bonds 123F
Adenine, bacteriophage MS2 RNA 340
Adenylate cyclase, G protein-mediated, activation 253 253F
Adenylate kinase 58F 59
ADP 115
ADP-vanadate 295
Alanine, collagen mutation 285 285F
Alanine, specificity of serine proteinases 213 214F
Alanine, structure 6F
Alanine, substrate-assisted catalysis 218
Alcohol dehydrogenase,  helix 18 helix 18
Alcohol dehydrogenase, helical wheel 17T
Alcohol dehydrogenase, zinc in 11 11F
Allosteric control 113 142
Allosteric effectors 142 (see also УRepressorФ)
Allosteric proteins, phosphofructokinase 114Ч117
Allosteric proteins, switch between T and R states 113Ч114 (see also УRepressorФ)
Allostery 113
Allostery, 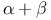 structures 32 structures 32
Allostery, 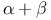 structures, Cro protein 132 structures, Cro protein 132
Allostery, 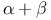 structures, lysozyme from T4 bacteriophage 355F structures, lysozyme from T4 bacteriophage 355F
Alphaviruses 340Ч341
Alphaviruses, catalytic triad in coat protein protease 341
Alphaviruses, core proteins 341 341F
AlzheimerТs amyloid  -protein precursor inhibitor (APPI) 361 362 362T -protein precursor inhibitor (APPI) 361 362 362T
AlzheimerТs disease 113 283 288
Amide-hydrogen exchange 95
Amino acid sequence 3F 4
|
|
 |
| –еклама |
 |
|
|
 |
|
ќ проекте
|
|
ќ проекте



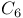 -zinc cluster family
-zinc cluster family  fragment
fragment 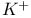 leak channels
leak channels 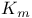 values
values 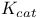 values
values 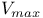
 domains
domains  structures, barrels
structures, barrels  motif
motif 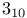 helix
helix 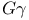

 strands
strands 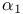 -Antitrypsin
-Antitrypsin 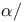 structure
structure  motifs
motifs  unit (hairpin
unit (hairpin 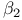 Microglobulin
Microglobulin  -Crystallin, coding sequence
-Crystallin, coding sequence 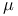 -oxo bridge
-oxo bridge 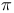 Helix
Helix 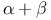 structures
structures