|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Kuchel P.W., Ralston G.B. — Schaum's outline of theory and problems of biochemistry |
|
|
 |
| Предметный указатель |
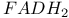 , in fatty acid oxidation 370 , in fatty acid oxidation 370
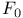 , of ATP synthase 412 , of ATP synthase 412
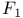 , ofATP synthase 412 , ofATP synthase 412
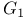 , phase of cell cycle 461 462d 463 , phase of cell cycle 461 462d 463
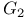 phase of cell cycle 461 462d 463 phase of cell cycle 461 462d 463
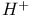 /ATP ratio, in oxidative phosphorylation 408d /ATP ratio, in oxidative phosphorylation 408d
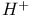 /phosphate symport, in mitochondria 409 /phosphate symport, in mitochondria 409
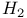 , in nitrogenase reaction 420d , in nitrogenase reaction 420d
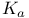 (acid dissociation constant), definition of 58 (acid dissociation constant), definition of 58
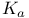 and and 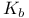 , relationship between 59 , relationship between 59
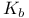 , basicity constant 59 , basicity constant 59
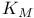 253d 256 257 261 262 262d 264d 268 276—278 281 282 284 285 287 288 289 253d 256 257 261 262 262d 264d 268 276—278 281 282 284 285 287 288 289
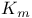 , expression for 260 , expression for 260
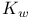 (ionic product of water) defined 57 (ionic product of water) defined 57
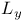 system, of amino acid transport 431 system, of amino acid transport 431
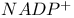 , NADPH in the pentose phosphate pathway 339d 340 , NADPH in the pentose phosphate pathway 339d 340
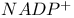 , NADPH, structure 312s , NADPH, structure 312s
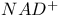 1 3-bisphosphoglycerate 1 3-bisphosphoglycerate
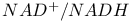 , conjugate redox pair 403 , conjugate redox pair 403
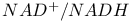 , hydride ion in 403 , hydride ion in 403
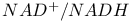 , light absorbance at 340 nm 403 , light absorbance at 340 nm 403
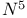 , , 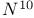 -Methenyltetrahydrofolate(5, 10, CH-THF) cyclohydrolase 442 -Methenyltetrahydrofolate(5, 10, CH-THF) cyclohydrolase 442
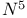 -Methyltetrahydrofolate 449 -Methyltetrahydrofolate 449
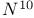 -Formyl tetrahydrofolate (10-CHO-THF) synthetase 442 -Formyl tetrahydrofolate (10-CHO-THF) synthetase 442
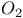 , transport in erythrocytes 15 , transport in erythrocytes 15
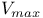 253 257 261 262d 264d 265d 268 276—278 281 282 284 285 287—289 253 257 261 262d 264d 265d 268 276—278 281 282 284 285 287—289
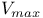 , expression for 260 , expression for 260
![$[1alpha]_D^T$](/math_tex/d533a5af6df905b992631ee5713e0a1e82.gif) 26 34 52 26 34 52
![$[^3H]$](/math_tex/65445266343aae3b0fb5144fa28ef38e82.gif) -Thymidine 4 -Thymidine 4
 linkage 47 linkage 47
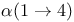 linkage 46 linkage 46
 linkage 46 linkage 46
 , ,  -Trehalose 52 -Trehalose 52
 , ,  -Trehalose 50s -Trehalose 50s
 -Actinin, in Z-disc 135 137 -Actinin, in Z-disc 135 137
 -Amanitin 494 500 -Amanitin 494 500
 -Cyano-3-hydroxycinnamate, inhibitor of pyruvate transport 184 -Cyano-3-hydroxycinnamate, inhibitor of pyruvate transport 184
 -D-Fructose 1, 6-bisphosphate 313s -D-Fructose 1, 6-bisphosphate 313s
 -D-Fructose 1, 6-bisphosphate in gluconeogenesis 325s -D-Fructose 1, 6-bisphosphate in gluconeogenesis 325s
 -D-Fructose 1, 6-bisphosphate in glycolysis 313s -D-Fructose 1, 6-bisphosphate in glycolysis 313s
 -D-Fructose 6-phosphate in gluconeogenesis 325s -D-Fructose 6-phosphate in gluconeogenesis 325s
 -D-Fructose 6-phosphate in glycolysis 313s -D-Fructose 6-phosphate in glycolysis 313s
 -D-Fructose 6-phosphate in the pentose phosphate pathway 341s -D-Fructose 6-phosphate in the pentose phosphate pathway 341s
 -D-Galactopyranose 38s -D-Galactopyranose 38s
 -D-Glucopyranosc 35 38 -D-Glucopyranosc 35 38
 -D-Glucose 6-phosphate in gluconeogensis 326 -D-Glucose 6-phosphate in gluconeogensis 326
 -D-Glucose 6-phosphate in hexokinase 313 -D-Glucose 6-phosphate in hexokinase 313
 -D-Glucose in gluconeogensis 326 -D-Glucose in gluconeogensis 326
 -D-Glucose in hexokinase 313 -D-Glucose in hexokinase 313
 -D-Mannopyranose 38s -D-Mannopyranose 38s
 -D-Methylglucopyranoside 44s -D-Methylglucopyranoside 44s
 -Galactosidase 383d -Galactosidase 383d
 -Glucosidase 11t -Glucosidase 11t
 -Helical tail, myosin 137 -Helical tail, myosin 137
 -Helical, intermediate filament 144d -Helical, intermediate filament 144d
 -Helices, in hemoglobin 117 -Helices, in hemoglobin 117
 -Helix, helix breaking amino acid residues 91t -Helix, helix breaking amino acid residues 91t
 -Helix, protein 112 -Helix, protein 112
 -Helix, spectrin 90 91d 150 151 -Helix, spectrin 90 91d 150 151
 -Hydroxy acid oxidase 12 -Hydroxy acid oxidase 12
 -Ketoglutarate, 2-oxoglutarate 420 -Ketoglutarate, 2-oxoglutarate 420
 -Lactalbumin, in lactose synthesis 115 -Lactalbumin, in lactose synthesis 115
 -Linolenic acid 156s -Linolenic acid 156s
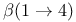 linkage, in cellulose 47 linkage, in cellulose 47
 -Alanine 56 70s -Alanine 56 70s
 -Carotene 163s 165 -Carotene 163s 165
 -Chains, hemoglobin 18 -Chains, hemoglobin 18
 -D-Methylfructofuranoside 44s -D-Methylfructofuranoside 44s
 -D-Ribofuranose 38s -D-Ribofuranose 38s
 -D-Ribopyranose 39s -D-Ribopyranose 39s
 -D-ribose 28s 49 51 198 -D-ribose 28s 49 51 198
 -Galactosidase 11f 383d -Galactosidase 11f 383d
 -Galactosidase in E. coli 508 -Galactosidase in E. coli 508
 -Globin gene 496 497d -Globin gene 496 497d
 -Glucosidase 11t 21 383d -Glucosidase 11t 21 383d
 -Glucuronidase 11t -Glucuronidase 11t
 -Glycosidic linkage, in nucleic acid 201 -Glycosidic linkage, in nucleic acid 201
 -Hydroxybutyrate dehydrogenase, location 13t -Hydroxybutyrate dehydrogenase, location 13t
 -Ketoacyl synthase 377t -Ketoacyl synthase 377t
 -Lactoglobulin 77 78 105 -Lactoglobulin 77 78 105
 -Methylgalactoside 51 -Methylgalactoside 51
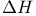 , enthalpy change 292 , enthalpy change 292
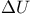 , internal energy change 290 292d , internal energy change 290 292d
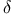 - Lactone 41s - Lactone 41s
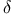 -Aminolevulinate synthase, ALA synthase 451 452 -Aminolevulinate synthase, ALA synthase 451 452
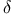 -Aminolevulinate synthase, ALA synthase, elF-2 in regulation 452 -Aminolevulinate synthase, ALA synthase, elF-2 in regulation 452
 -Aminolevulinate synthase, ALA synthase, inhibition by hemin 452 -Aminolevulinate synthase, ALA synthase, inhibition by hemin 452
 -Aminolevulinate synthase, ALA synthase, protein kinase and 452 -Aminolevulinate synthase, ALA synthase, protein kinase and 452
 -Aminolevulinate synthase, ALA synthase, regulation 452 -Aminolevulinate synthase, ALA synthase, regulation 452
 -Aminolevulinate, -Aminolevulinate, 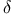 -aminolevulinic acid, (ALA) 451s -aminolevulinic acid, (ALA) 451s
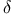 -Gluconolactone 41 ; -Gluconolactone 41 ;
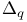 , heat change 292 , heat change 292
 -Aminobutyrate (GABA) 70s 455 -Aminobutyrate (GABA) 70s 455
 -L-Glutamyl-L-cysteine 240d -L-Glutamyl-L-cysteine 240d
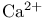 , Calcium ions in muscle 17 , Calcium ions in muscle 17
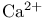 , Calcium ions, effect of ions on soaps 183d , Calcium ions, effect of ions on soaps 183d
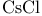 , in density gradient centrifugation 7 , in density gradient centrifugation 7
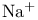 symport 430 symport 430
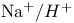 antiport, in mitochondria 409 antiport, in mitochondria 409
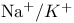 ATPase 430 ATPase 430
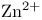 as electrophile 249 as electrophile 249
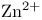 in carbonic anhydrase 229 in carbonic anhydrase 229
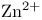 in carboxypeptidase A 248 in carboxypeptidase A 248
 , genome size 215 216t , genome size 215 216t
 , single-stranded circular DNA 226 , single-stranded circular DNA 226
 , viral DNA 510 , viral DNA 510
1, 2, 3-Triacylglycerol 378d
1, 2, 3-Triacylglycerol, synthesis of 380d
1, 2-Diacylglycerol 378d 380d 382d
1,3-Bisphosphoglycerate, in substrate level phosphorylation 402s
10-Methylstearic acid 157s
2', 3'-ddCTP 486
2', 3'-Dideoxynucleosides 473
2, 3-Bisphosphoglycerate 344
2, 3-Bisphosphoglycerate, Rapoport — Luebering shunt and 344
2, 3-Bisphosphoglycerate, synthesis 344
2, 3-Oxidosqualene: lanosterol synthase 391d
2-Amino-2-deoxy-D-glucose, D-glucosamine 42
2-Deoxy- -D-ribose 39, 40s 201s -D-ribose 39, 40s 201s
2-Hydroxybenzoic acid 398s
2-Monoacylglycerol 362s 378d
2-Oxoacid 423t
2-Oxoacid dehydrogenase (BCOADH) 432
2-Oxoacid dehydrogenase (BCOADH), location 13t
2-Oxoglutarate 240s 422s
2-Oxoglutarate dehydrogenase complex, product inhibition by NADH and succinyl-CoA 351
2-Oxoglutarate dehydrogenase complex, reaction 347s
2-Oxoglutarate in the citric acid cycle 347s
2-Oxoglutarate,  -ketoglutarate 420 -ketoglutarate 420
2-Oxoglutarate, product of prolylhydroxylase 123d
2-Phosphoglycerate, in glycolysis 315s
2-Phosphoglycerol 3d 4 11t
2-Thenoyltrifluoroacetone, inhibition of electron transport chain 406d
20, 22-Dihydroxycholesterol 393d
22-Hydroxycholesterol 393d
3', 5'-AMP, cAMP 227
3, 4-Dihydroxyphenylalanine (dopa) 432s
3-Hydroxy-3-methylglutaryl-CoA 388 389s
3-Hydroxyacyl-CoA dehydrogenase 372t
3-Hydroxybutyrate dehydrogenase,  -hydroxybutyrate dehydrogenase 373t -hydroxybutyrate dehydrogenase 373t
3-Hydroxybutyrate,  -hydroxybutyrate 37t 370s 371 394 396 -hydroxybutyrate 37t 370s 371 394 396
3-Oxoacid transferase 374
3-Phosphoglycerate 425d
3-Phosphoglycerate in glycolysis 315s
3-Phosphoglycerate in substrate level phosphorylation 402s
3-Phosphohydroxypyruvate 425d
3-Phosphoserine 425d
4-Methylpentanal 3934
5'  3'Exonuclease 465 466 3'Exonuclease 465 466
5'  3'Exonuclease, activity of DNA polymerase I 466 3'Exonuclease, activity of DNA polymerase I 466
| 5-Bromouracil 199s 200s
5-Bromouracil, 2,3-dideoxy- -D-ribose 220s -D-ribose 220s
5-Fluorouracil 219 220s 444s 445d
5-Hydroperoxyeicosatetraenoic acid 387d
5-Lipoxygenase 3874
5-methylcytosine 199s
5-Phosphoribosyl, 1-pyrophosphate (P-Rib-PP) 437 438 440d 440s
5S RNA 494
6-Aminopurine 200
6-Meracaptopurine 444s 445d
6-Oxy-2-aminopurine 200
6-Phosphogluconate, in the pentose phosphate pathway 339s
6-Phosphogluconolactone, in the pentose phosphate pathway 339s
7- -Hydroxycholesterol 391d -Hydroxycholesterol 391d
Abietic acid 185s
Absorption, of dietary lipid 391
Absorptive cells 17
ACAT, acyl-CoA: cholesterol acyltransferase 366d 390
Acetal 32s 43s
Acetaldehyde 232s 322s
Acetate, as precursor of lipids 154
Acetic acid as pH buffer 72s
Acetic acid, oxidation energetics 304
Acetic acid, pKa 59
Acetoacetate 310s 373sd 396
Acetoacetate, utilization 374d
Acetoacetyl-CoA 374 388s
Acetogenins, acetate as precursor 154s
Acetone 370s 373sd
Acetyl phosphate, energetics of 309d
Acetyl transacylase 377t
Acetyl-CoA 370 374 374d 388s 396
Acetyl-CoA carboxylase 375 394
Acetyl-CoA in the citric acid cycle 345s 346s 350s
Acetyl-CoA in the glyoxylase cycle 356s
Acetyl-CoA, acetyltransferase 373t
Acetyl-CoA, sources in metabolism 351
Acetylcysteinyl-polypeptide 376d
Acid anhydride bond 231t
Acid catalysis 233
Acid deoxyribonuclease 11t
Acid hydrolase 9 10d
Acid hydrolases, in lysosomes 20d
Acid phosphatase 3d 11t
Acid phosphodiesterase 11t
Acid ribonuclease 11t
Acid, acid-base behavior 58 59
Acid, Bronsted definition of 58
Acid, conjugate acid 58 59
Acid, weak acids 58 59
Acid-base catalysis, general 232 233
Acidfuchsin 19
Acidophilic, stain 19
Aconitase 13t 346 347d
Aconitase, iron-sulfur protein 347
Aconitase, location 13t
Acridine 473 474s
acrosomal process 108
Actin 113ct 132 133 139 150
Actin double helix 113
Actin filament 12
Actin, (minus) end 133d
Actin, (plus) end 133d
Actin, ADP with 132
Actin, assembly 132d 134
Actin, ATP with 132
Actin, barbed end 133 134d
Actin, capping proteins and 135
Actin, critical concentration 132 133 134d
Actin, double helix of 113 131 132
Actin, F- 134d
Actin, G- 134d
Actin, lag period in polymerization 132
Actin, nucleus 132d
Actin, pointed end 133 134d
Actin, polarity 133
Actin, polymer 131
Actin, polymerization 132d
Actin, thymosin binding 134
Actin, treadmill 133
Actin, various forms 136d
Actin-binding protein 134 135 140 150
Actinomycin D 473 499 500 513
Activation energy 234 237 247
Activation of amino acids, for protein synthesis 501d
Active site 238
Active site of enzyme 233 234
Active transport, across membranes 177 178t
Active transport, examples 179d
Active transport, group translocation 179d
Active transport, primary active 179d
Active transport, secondary active 179d
Activity coefficient 251
Activity coefficient, definition 305d
Actomyosin 17 144
Actomyosin, conformational change 139d
Actomyosin, power stroke 138
Acyl transacylase 377t
Acyl-CoA dehydrogenase 370 372t
Acyl-CoA derivatives 370
Acyl-CoA desaturase 397
Acyl-CoA synthetase 369 378
Acyl-CoA transferase 378
Acyl-CoA: cholesterol acyltransfer (ACAT) 366d 390
Acyl-enzyme intermediate 241d 242d
Acylate, fatty acid salt 155
Acylimidazole 242
Adair equation 268 269 271 289
Adair, G, S, 268
Adamantane 245s
Adenine 200s 205s 458
Adenosine 201s 202
Adenosine 5'-phosphate, AMP 202s 203
Adenosine deaminase 456
Adenosine triphosphate, ATP 203
Adenyl cyclase, in glycogenosis 337d
Adenylate cyclase 392
Adenylate kinase 436
Adenylate kinase, location 13t
Adenylate kinase, thermodynamics of 300d
Adenylic acid, AMP 203
Adiabatic (closed) system, definition 290 291
Adiabatic calorimeter 291d 304
Adipocyte 16d 17
Adipose tissue 392
ADP in glycolysis 311
ADP in muscle contraction 137 138 139d
ADP-3'-phosphate, in CoA 345s 346
ADP-ribosylation 508
Adrenal cortex 336
AG, change in Gibbs free energy 293
Aglycone 43
Alanine 17 67s 238 423t
Alanine racemase 241
Alanine transaminase 241
Alanine transaminase, aminotransferase 241
Alanine transaminase, pyridoxal phosphate in 241
Alanine, aminotransferase 422d
Alanine, structure of 54t
Alcaligenesfaecalis 227
Alcohol dehydrogenase 249 322d
Alcohol dehydrogenase, horse-liver 289
Aldehyde 3
Aldehyde, groups 25 42 43
Alditol 40
Aldohexose 25 28
Aldol 125d
Aldolase in glycolysis 314d
Aldolase, molecular weight 83t
Aldonate 41
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



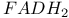 , in fatty acid oxidation
, in fatty acid oxidation 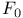 , of ATP synthase
, of ATP synthase 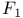 , ofATP synthase
, ofATP synthase 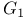 , phase of cell cycle
, phase of cell cycle 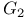 phase of cell cycle
phase of cell cycle 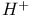 /ATP ratio, in oxidative phosphorylation
/ATP ratio, in oxidative phosphorylation 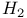 , in nitrogenase reaction
, in nitrogenase reaction 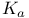 (acid dissociation constant), definition of
(acid dissociation constant), definition of 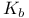 , relationship between
, relationship between 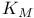
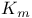 , expression for
, expression for 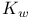 (ionic product of water) defined
(ionic product of water) defined 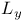 system, of amino acid transport
system, of amino acid transport 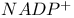 , NADPH in the pentose phosphate pathway
, NADPH in the pentose phosphate pathway 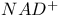
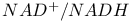 , conjugate redox pair
, conjugate redox pair 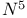 ,
, 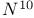 -Methenyltetrahydrofolate(5, 10, CH-THF) cyclohydrolase
-Methenyltetrahydrofolate(5, 10, CH-THF) cyclohydrolase 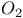 , transport in erythrocytes
, transport in erythrocytes 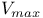
![$[1alpha]_D^T$](/math_tex/d533a5af6df905b992631ee5713e0a1e82.gif)
![$[^3H]$](/math_tex/65445266343aae3b0fb5144fa28ef38e82.gif) -Thymidine
-Thymidine  linkage
linkage 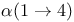 linkage
linkage  linkage
linkage  ,
,  -Trehalose
-Trehalose 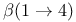 linkage, in cellulose
linkage, in cellulose 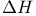 , enthalpy change
, enthalpy change 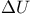 , internal energy change
, internal energy change 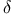 - Lactone
- Lactone 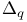 , heat change
, heat change  -Aminobutyrate (GABA)
-Aminobutyrate (GABA) 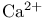 , Calcium ions in muscle
, Calcium ions in muscle 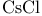 , in density gradient centrifugation
, in density gradient centrifugation 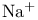 symport
symport 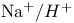 antiport, in mitochondria
antiport, in mitochondria 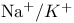 ATPase
ATPase 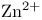 as electrophile
as electrophile  , genome size
, genome size  3'Exonuclease
3'Exonuclease