|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Kuchel P.W., Ralston G.B. — Schaum's outline of theory and problems of biochemistry |
|
|
 |
| Предметный указатель |
Aldonic acid 41
Aldopentose 26 28
Aldose 25
Algae, blue-green 419
Allolactose 508
Allose 28s
Allosteric, coefficient 273d
Allosteric, effect 270
Allosteric, effector site 261
Allosteric, interactions in hemoglobin 118
Allysine 124 125d 126
Altman, C 258
Altmann, R. 12
Altrose 28s
Amanita phalloides 494
Amidino 230s
Amine oxidase 434
Amino acid 17 23 53s—75
Amino acid as zwitterion 61—65
Amino acid with prototropic side chains 63
Amino acid, acid-base behavior 56—65
Amino acid, activation 501d
Amino acid, analysis 65
Amino acid, essential 419
Amino acid, fates of carbon skeleton 431d
Amino acid, formal titration of 62
Amino acid, isoelectric point 62 64 65
Amino acid, lifetime in plasma 431
Amino acid, ninhydrin reaction with 65s
Amino acid, pKa values 63f
Amino acid, plasma concentration 431
Amino acid, reaction with ninhydrin 65
Amino acid, site of synthesis 17
Amino acid, structure of 53s
Amino acid, synthesis 419
Amino acid, transport 430
Amino acid, tritiated 4
Amino sugars 42
Aminoacyl-tRNA 504d 507s
Aminoacyl-tRNA synthetase 501d 514 518
Aminopeptidase 248 430 505
Aminotransferases 421 422 423d
Ammonia 419 420
Ammonia, equilibrium with ammonium 434d 435
AMP 240d
AMP, adenosine 5'-phosphate 202s 203s
AMP, binding to phosphorylase a 115
Amphibolic reactions, citric acid cycle 354 355t
Amphipathic 165
Amphiphilic lipids 165 167
Amplification in PCR 480
Amplification of DNA 464d
Amplification of metabolic signals 301
Amylo (1, 4 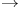 6)-transglycosylase, branching enzyme 328d 6)-transglycosylase, branching enzyme 328d
Amylopectin 46
Amylopectin, structure 330
Amylose 46 51
Amylose, structure 330
Anaphase 142 143 144
Anaplerotic reaction of the citric acid cycle 355t
Anaplerotic reaction, C1 compounds 449
Andrews, PR, 244 245
Androgen 393d
Anemia 18 131
Anhydride(s) 242 243
Animal 1
Anion channel 136 137d
Anion transport, in erythrocytes 16
Ankyrin 136 137d
annealing 213
Annealing, DNA 477d 479 480d
Anticodon 500 501d
Anticompetitive inhibition 208d 254 255t 264
Antilipolytic hormone, insulin 394
Antimicrobial agents 245
Antimycin A, inhibition of electron, transport chain 406d
Antisense strand, template 489
Aphidicolin 474 475s
Apo proteins of lipoproteins, AI, AII, B, CI, CII, CIII, D 169
Apoferritin 453
Apolipoprotein B 366
Apolipoprotein C 365
Apolipoprotein E 366
Apoprotein 81
Arabino-hexulose, D- (fructose) 30 31
Arabinose 28s 47
Arachidonic acid 156s 385sd 386d 387d 398
Arachidonic acid, arachidonate 376 377 384 385s
Arachidonic acid, synthesis of 377d
Arginine 230s 238 423t 424 430s 436s 436
Arginine, amidinohydrolase 230
Arginine, structure of 56t
Argininosuccinate 436s
Argininosuccinate lyase, argininosuccinase 249 424 436d
Argininosuccinate synthetase 424 436
Arginyl, staining 2
Aromatic compound 2
Arrhenius, S, , activation energy 237 238
Arthritis 129
Arylhydrazinopyrimidine 474
Arylsulfatase 11t 23
Arylsulfatase, A 383d
AS, entropy change 292
Ascorbate 505
Ascorbate and prolylhydroxylase 123d
Ascorbate as secondary redox mediator 406
Asparagine 70s 423t
Asparagine, structure of 54t
Aspartate 70s 283 422s 423t
Aspartate carbamoyltransferase 229 266
Aspartate carbamoyltransferase, active site 266
Aspartate carbamoyltransferase, allosteric site 266
Aspartate carbamoyltransferase, aspartate as substrate 229
Aspartate carbamoyltransferase, ATP and 266
Aspartate carbamoyltransferase, ATP binding 115
Aspartate carbamoyltransferase, carbamoyl aspartate in 229
Aspartate carbamoyltransferase, carbamoyl phosphate as substrate 229
Aspartate carbamoyltransferase, catalytic subunit 115
Aspartate carbamoyltransferase, CTP and 266
Aspartate carbamoyltransferase, CTP binding 115
Aspartate carbamoyltransferase, effector molecules 229 266
Aspartate carbamoyltransferase, Escherichia coli (E. coli) form 266
Aspartate carbamoyltransferase, inhibitor(s) 229
Aspartate carbamoyltransferase, subunit structure 115 116d
Aspartate carbamoyltransferase, subunits of 229
Aspartate carbamoyltransferase, X-ray diffraction of 115
Aspartate semialdehyde 283
Aspartate, aminotransferase 422
Aspartate, structure of 55t
Aspartic acid, ionization of 71s
Aspartic acid, isoelectric point of 71
Aspartokinase 283
Aspirin 398s
Association binding constant 268
Astral microtubule 143d
Atmospheric nitrogen 419
ATP 9 21 240d
ATP and actomyosin 139
ATP and hexokinase 237
ATP in glycolysis 311
ATP in muscle contraction 137—139d
ATP in pyruvate carboxylase reaction 353d
ATP synthase 13t 402
ATP synthase in bacteria 411
ATP synthase in chloroplasts 411
ATP synthase in mitochondria 411
ATP synthase, energetics of 309
ATP synthase, location 13t
ATP synthase, mechanism 413d 414d
ATP synthase, subunit structure 412
| ATP translocase 241
ATP translocase in mitochondria 415
ATP translocase, inhibitors 415
ATP utilization in macrophages 21
ATP, adenosine triphosphate 203
ATP, bioenergetic properties 298
ATP, free energy hydrolysis 402
ATP, production in glycolysis 314
ATP, structure 203s
ATPase 237s 289
ATPase in muscle contraction 137—139d
Atractyloside, inhibitor of ATP, translocase 415
Attachment glycoproteins 130
Attenuator sequence, mRNA 512
Autonomously replicating sequences (ars) 472
Autophagic vacuole 19 20d
Autophagosome 20d
Autophagy 9 20d
Autoradiography 4 23
Autosomes 15
Average life span of a cell 21
Avogadro’s number 22
Axial configuration 36s
Axon 140 141d
Axoneme, in cilia 141 142d
Azide, inhibition of electron transport chain 406d
Azure I 1
bacteria 12 21 419
Bacteria, bacterium 1
Bacteria, phagocytosis of 21
Bacterial cell wall 11t
Bacterial cell wall, polysaccharides 245
Bacterial plasmid 477
Bacteriophage T4, virus 110d 111d
Bacteriophage T5, tobacco mosaic 110
Bacteriophage T6, polio 110
Bacteriophage, genome size 215 216t
Barbed end of actin 136
Barbed end,  -actinin binding to 137 -actinin binding to 137
Basal body 142
Base catalyst 233
Base pairs in genes 149
Base, definition of 59
Base-pairing geometry 210
Basement membrane 122t
Basement membrane, binding 131t
Basement membrane, structure of 125d
Basic fuchsin 2s
Basophilic substance 18
BCOADH, branched chain 2-oxoacid dehydrogenase 432
beans 419
Becker muscular dystrophy 131
Beer — Lambert law, equation in studies of electron transport chain 415d
Beer — Lambert law, law, relationship 69
Beer — Lambert law, relationship 53 56
Bell-shaped curve 262d
Benzene, structure 153s
Bi Bi enzymatic reaction 259d 289
Bicarbonate in pyruvate carboxylase reaction 353d
Bicarbonate, dehydration 228
Biconcave disk, shape 15 16d
Bidirectional replication 460
Bifunctional enzyme 438 440
Bilayer, lipid 167d
Bile, acid definition 168s
Bile, cannaliculus 17
Bile, definition 168s
Bile, micelle 363d
Bile, salt 387 390 395
Bile, synthesis from cholesterol 391d
Bilirubin 453s
Bilirubin, diglucuronide 453
Bilirubin, sulfate 453
Binding energy 234
Binding function, Adair 269
Binding proteins 267
Bioblast 12
Biochemical standard state 293
Bioenergetics, definition 402
Biopterin 455
Biosynthesis of purine nucleotides 440
Biotin 374s
Biotinylenzyme 374d
Bird 455
Blood group substance 16
Blood vessel 17
Blue-green algae 419
Boat 36 37s
Bohr, C 267
Boltzmann, L, 238
Bond, angle 234
Bond, length 234
Bond, strain 234
bone 11t 17 121t 122t
Bone, collagen 148
Bongkrekic acid, inhibitor of ATP translocase 415
Boric acid, pKa 59
Bottlebrush structure 127 129
BPG as an allosteric effector 120
BPG, 2, 3-bisphosphoglycerate 119s 120 121 147
BPG, binding to hemoglobin 120 151
BPG, competition with 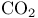 120 120
BPG, low affinity for oxyhemoglobin 120
BPG, synthesis from glucose 120
Brain 11t 21
Brain, lipids 362
Branched metabolic pathways 265d
Branching enzyme, amylo-( 1, 4 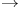 1, 6)-transglycosylase 328d 1, 6)-transglycosylase 328d
Briggs and Haldane 256
Briggs and Haldane, analysis 263d
Briggs, G, E, 256
Bronsted — Lowry, definition of acid and base 58 233
Brown fat, uncoupled oxidative phosphorylation 417
Brush border 136
Buffer, amino acids as 61—65
Buffer, pH 59
Bulk properties of matter 290
Butyrylcysteinyl-polypeptide 376d
Calorimeter 291d
Camitine-fatty acid acyltransferase, location 13t
Camosine 68s
cAMP dependent protein kinase 399d
Camphor 185s
Cancer 458
Cannaliculus, bile 17
Cap-binding proteins 505
Capping of RNA 497
Carbamate, of hemoglobin 121s
Carbamoyl phosphate 435s
Carbamoyl phosphate synthetase 1, ammonia-dependent 438
Carbamoyl phosphate synthetase 11 435
Carbamoyl phosphate synthetase 11, glutamine-dependent 438
Carbamoyl phosphate synthetase, compartmentation of 24
Carbamoyl phosphate synthetase, location 13t
Carbocation 3 247
Carbohydrate 2 21 25
Carbohydrate, classification 25
Carbohydrate, dietary 374
Carbon dioxide production, citric acid cycle 249d 348 357
Carbon dioxide production, pentose phosphate pathway 340s
Carbon dioxide production, pyruvate dehydrogenase 352d
Carbon dioxide,  15 15
Carbon dioxide, 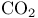 , binding to hemoglobin 121 , binding to hemoglobin 121
Carbon dioxide, 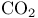 , hydration 228 , hydration 228
Carbon monoxide, inhibition of electron, transport chain 406d
Carbonic acid 228
Carbonic anhydrase 228
Carbonic anhydrase, 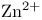 in 229 in 229
Carbonium ion 247
Carboxypeptidase 427 428d
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



 6)-transglycosylase, branching enzyme
6)-transglycosylase, branching enzyme  -actinin binding to
-actinin binding to 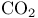
 in
in