|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Kuchel P.W., Ralston G.B. — Schaum's outline of theory and problems of biochemistry |
|
|
 |
| Предметный указатель |
Hexadecanoyl-CoA 369s
Hexagonal close-packing, of spheres 113
Hexokinase 232d 236 237 276 344
Hexokinase, 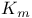 for glucose 317 318d for glucose 317 318d
Hexokinase, ATP as substrate 236
Hexokinase, binding energy of 237
Hexokinase, first step in glycolysis 313d
Hexokinase, glucose 6-phosphate product 237s
Hexokinase, glucose as substrate 236s
Hexokinase, induced fit in 237
Hexokinase, nucleophilic attack in 237
Hexokinase, product inhibition of 317
Hexokinase, substrates 237
Hexosaminidase A and B 383d 397
Hexose 25
Hexose monophosphate 339
Hierarchy of structure in cells 17
High-density lipoprotein (HDL), definition 168 169t 364 390
High-lysine corn 419
Hill coefficient 268 287
Hill coefficient in hemoglobin 148 151
Hill equation 267 286d 289
Hill plot 268
Hill, A, V. 151 267 268
Histamine 21 22sd 240s 455
Histamine in mast cells 21
Histidine 233 423t
Histidine as pH buffer 72
Histidine decarboxylase 241
Histidine, structure of 56t
Histidyl 231
Histidyl, staining 2
Histochemistry 3 23
Histology 2 18
Histone 14 216 473
Histone, genes for 498
Histone, H1 217d
Histone, H2A 217d
Histone, H2B 217d
Histone, H3 217d
Histone, H4 217d
HMG-CoA lyase 373t
HMG-CoA reductase 388d 394 399
HMG-CoA synthesis, kinase 399d
hnRNA, heterogeneous nuclearRNA 497
Hogness box, TATA box 494 496
Holland 1
Homo- -linolenic acid 398 -linolenic acid 398
Homocysteine 449s
Homocysteine, transmethylase 449
Homogenate 4
Homogenization 5
Homogenizer 19
Homologous chromosomes 14
Homolytic bond cleavage 245
Homoserine dehydrogenase 283
Homotropic effect 266 267 271 273d 270
Hormone-sensitive lipase 368 392 394
Horse-liver alcohol dehydrogenase 289
Human, cytochrome c 98td
Hyaloplasm 6d
Hyaluronan 127d 128d 129
Hyaluronan, core protein 126
Hyaluronic acid 11t 47
Hyaluronidase 11t 129
Hydride ion 403
Hydrogen bond 86t 247
Hydrogen bond between bile salt molecules 168
Hydrogen bond in cellulose 47
Hydrogen bond in collagen 123
Hydrogen bond in hemoglobin 148
Hydrogen bond with amino acids 75
Hydrogen electrode 297d
Hydrogen peroxide, formation 12
Hydrogen, in nitrogenase reaction 420d
Hydrolase 229 230t
Hydrolase, classification 231t
Hydrolyic cleavage 230t
hydrolysis 231d
Hydrolysis reaction 233 234
Hydrolytic enzyme 16 21
Hydrolytic enzyme in erythrocytes 16
Hydronium ion 57d
Hydrophobic effect, with lipids 166
Hydrophobic interaction 86t 123 234
Hydrophobic interaction in elastin 126
Hydrophobic interaction in spectrin 150
Hydrophobic pocket 249
Hydrophobic residues, in collagen 148
Hydroxyacyl dehydratase 377t
Hydroxyacyl-CoA dehydrogenase 370
Hydroxyapatite 121
Hydroxyisovalerate, in valinomycin 181
Hydroxylysine 505
Hydroxymethylglutaryl-CoA (HMG-CoA) synthase 373t
Hydroxymethylglutaryl-CoA reductase 388d 389d
Hydroxyproline 70s 93s 123d 505
Hydroxyproline in collagen 93 148
Hydroxyurea 444s
Hyperammonemia 455
Hyperchromic effect, in single-stranded DNA 212
Hypoxanthine 447d
Ideality, thermodynamic 114
Idose 28s
Imidazole 75s
Immunodeficiency 456
IMP, inosine 5'-monophosphate 225s
Inborn errors of metabolism 15
Induced fit hypothesis 236 273
Inducer, of gene 508
Infant 15
Inflammatory white cells 1
Influenza virus, hemagglutinin 112 113d
Inherited disease 21
Inhibition of enzyme, anticompetitive 254
Inhibition of enzyme, competitive 254
Inhibition of enzyme, mixed 254
Inhibition of enzyme, noncompetitive 254
Inhibition of enzyme, uncompetitive 254
Inhibition pattern 280
Inhibitor 253
Initial reaction rate 274
Initial velocity 252
Initiation codon 490t
Initiation codon, AUG 510
Initiation, of DNA replication 460 469
Inner sheath, in cilia 141 142d
Inosine 218s 447d
Inosine 5'-monophosphate (IMP) 225s
Inositol, in phosphatidylinositol 159t
Insulin 394
Insulin, affect on glycolysis 336
Insulin, gene 518
Insulin, self association 111 112
Insulin, structure 104 506d
Integral protein, in membranes 172
Integrin 130
interaction energy 234
Intercalation into DNA 473
Interconvertible enzyme 301 302
Intermediate density lipoproteins (IDL) 364
Intermediate density lipoproteins (IDL), definition 168 169t
Intermediate filament 12 131 144 145t 151
Internal energy change (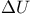 ) 290 292 ) 290 292
International Union of Biochemistry and Molecular Biology (IUBMB) 241
International Union of Pure and Applied Chemistry (IUPAC) 241
Interphase 140
Intervertebral disk 122t 127 129
Intestinal epithelial cell 363d
Intestine 17
| Intestine, secretory cell in 17 18d
Intracellular 21
Intracellular vesicle, size 17
Intracellular, compartmentation 303
Intracellular, symbionts 12
Intranucleolar chromatin 14d
Intrinsic association binding constant 271
Intrinsic binding constant 287
Intrinsic binding constant, definition 269
Intrinsic protein, in membranes 172
Intron 487 497
Inuit, Eskimo 419
Invertase 52 255
Iodate 3
Iodine in, oligonucleotide synthesis 211
Iodoacetate, modification of cysteine by 69s
Ion-exchange chromatography 77
Ion-exchange chromatography in amino acid analysis of proteins 65 66d
Ionic strength 299 305d
Ionizing radiation 475
Ionophore 181
Iron-sulfur proteintcluster in electron transfer chain 405t
Iron-sulfur proteintcluster, aconitase 347
Iso mechanism, enzymatic reaction 259
Isocitrate 347s
Isocitrate dehydrogenase 13t
Isocitrate dehydrogenase, activation by ADP and 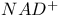 351 d 351 d
Isocitrate dehydrogenase, inhibition by ATP and NADH 351d
Isocitrate dehydrogenase, reaction 347d
Isocitrate in the glyoxylase cycle 356s
Isocitrate lyase 356d
Isoelectric point, glutamic acid 64t
Isoenzyme 321
Isolated (adiabatic) system, definition 290
Isolation of genes 476
Isoleucine 283 423t
Isoleucine, structure of 54t
Isomerase 230t
Isomerization, enzyme 259
Isopentenyl pyrophosphate 388s 389d 390d
Isoprene 387
Isoprene, acetate as precursor 154
Isopycnic centrifugation 5d
Isozyme 285
IUBMB, International Union of Biochemistry and Molecular Biology 241
IUPAC, International Union of Pure and Applied Chemistry 241
Jansen, E.F. 239
Janus green B 12
Joints 17
Karyotype 15
Katal 257
Keratan sulfate 127 128d
Keratin, attachment to fibroblasts 145
Ketimine 242s
Ketoacyl reductase 377t
Ketogenesis 370s 371 373d
Ketoheptose 26
Ketone body 370s 371
Ketone body, metabolism 394d
Ketone body, utilization in starvation 394d
Ketone group 42
Ketopentose 26
Ketose 25 30
Ketotriose 26
Kidney 11t
kinesin 141d 144
Kinesin in cellular motors 143
Kinetic order 274
Kinetic stability 237
Kinetics of membrane transport 175d
Kinetochore 143d 144
King and Airman, procedure 257 258 260d
King, E.L. 258
Klenow fragment, of DNA polymerase I 466
KNF model, Koshland, Nemethy and Filmer 273
Koshland, D, E. 273
Krabbe leukodystrophy 383d
Krebs cycle, citric acid cycle, tricarboxylic acid 345 346
Krebs, H, A., urea cycle discovery 435
Kynurenine hydroxylase 13t
Kynurenine hydroxylase, location 13t
L- Xylulose 31s
L-Alanine 240s
L-Amino acid oxidase 12
L-Fucose 40s
L-Glutamate 240
L-Histidine 240s
L-Iduronic acid 52s
L-Omithine 435d
L-Rhamnose 40s
L-Ribitol 1 -phosphate 49 50s
L-Ribulose 5-phosphate 240d
L-Threo-pentulose 30
L-Threose 28
L-Tyrosine 240d
L-Xylulose 5-phosphate 341s
Lac operon, in ?, coli 516
Lactase 330 343
Lactase, conversion to pyruvate 321s
Lactase, redox recycling 322d
Lactate dehydrogenase 249
Lactate dehydrogenase in Cori cycle 327d
Lactate dehydrogenase, estimation of 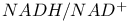 ratio 303d 304 ratio 303d 304
Lactate dehydrogenase, in 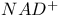 recycling 321d recycling 321d
Lactate dehydrogenase, isoenzymes of 327
Lactate fermentation 321
Lactate oxidase 12
Lactobacillic acid 182s
Lactonase 339d
Lactose 50s 330
Lactose (LAC) operon 508d
Lactose (LAC) operon in E. coli 508
Lactose (LAC) operon, catabolic activator protein 508
Lactose (LAC) operon, lac operon 508
Lactose synthetase 115
Lactose, synthesis 115
Lagging 466d
Lagging, strand 457 469d
Laminin 131t
Lamins 151
Lamins in prophase 145
Lanosterol 389 391d
LCAT, lecithin, cholesterol, acyltransferase 3654
LDL receptor 366d 368 390
LDL, low-density lipoprotein definition 168 169t
Lead phosphate 4
Lead zirconate titanate, piezoelectric effect of 19
Leading strand 467 4694
Leeuwenhoek, Antonie van 1
Lennard — Jones 6, 12 potential 85
Lesch — Nyhan syndrome 446
Leucine 423t
Leucine, structure of 54t
Leukemia, treatment 444
Leukotriene, 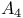 387d 387d
Leukotriene, 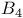 3874 3874
Leukotriene, synthesis from arachidonic acid 387d
Ligament, 121f 125 149
Ligand 270
Ligand, binding 115
Ligase 230t 241
Light micrograph 4 23
Light micrograph of cells 23
Lignoceric acid 161
Limonene 162s
Limonene, structure 153s
Lineweaver — Burk plot 253 254d 264d 265 266d 271 278 279 285f 286d
Link protein, in proteoglycan 127 128d
Linoleic acid, 155s 156 265 365 377d 395
Linoleic acid, linoleate 376 377
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



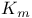 for glucose
for glucose  -linolenic acid
-linolenic acid 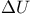 )
)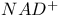
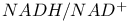 ratio
ratio