|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Kuchel P.W., Ralston G.B. — Schaum's outline of theory and problems of biochemistry |
|
|
 |
| Предметный указатель |
NADPH, in fatty acid synthesis 376 377r
Nail, finger 145
Nalidix acid 474 475s
Nasal lining, cilia in 17
Nascent fragments 466
Nebulin, and vernier principle 138
Negative control of a gene 508
Negative cooperativity 269 284 285
Negative cooperativity, enzyme kinetics and binding 266
Negative effector, of aspartate carbamoyltransferase 229
Negative feedback inhibition in the citric acid cycle, succinate dehydrogenase 351
Negative feedback inhibition of enzyme 265d 283d
Negative feedforward control 284
Negative nitrogen balance 419
Negative supercoiling of DNA 468
Nemethy 273
Nemst potential equation 307
Nemst redox equation 297d 406d
Nerve gas, diisopropyl fluorophosphate 239s
Nerve, squid 1
Nervonic acid 161
Neurofilaments 145t
Neurospora, cytochrome c 98t4
Neurotransmitter catabolism, location 13t
Nexin, in cilia 141 142d
Nick in DNA 464
Nick translation of DNA 467
Nickel, in urease 241
Nicotinamide adenine dinucleotide, (NAD) 311s 312
Niemann-Pick disease 382t 383d
Ninhydnn, reaction with amino acids 65s
Nitramide 233s
Nitrite 418
Nitrogen, atmospheric 419
Nitrogen, balance 419
Nitrogen, fixation 419
Nitrogen, fixing bacteria 420
Nitrogenase 4194
Nitrogenase, 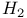 in 4204 in 4204
Nitrogenase,  419 419
Nitrogenase, ATP in 4204
Nitrogenase, flavoprotein in 420
NMR (nuclear magnetic resonance spectroscopy) 100
NMR (nuclear magnetic resonance spectroscopy) of protein 239
Nobel prize 1975 13
Nojirimycin 52
Nomenclature 383
Noncompetitive inhibition 254 255t 264 280d 289
Noncompetitive inhibition of enzyme 263d
Nonequilibrium reaction 299
Nonessential amino acid 423t
Nonhomologous recombination of DNA 463
Nonideality 305
Nonideality, thermodynamic 114
Noninteracting binding sites 269
Nonlinear least squares regression 254
Nonprotein amino acid 56
Nonsense codons 491
Norepinephrine 432 433s
Novobiocin 474
Nuclear envelope 7 144 142 151
Nuclear lamin 145t
Nuclear lamina, composed of lamin 145
Nuclear magnetic resonance, spectroscopy (NMF), of protein 239
Nuclear magnetic resonance, spectroscopy, NMR 100
Nuclear membrane 64 144
Nuclear pore 144
Nuclease 11t 219
Nucleic acid 198
Nucleic acid, breakdown 4474
Nucleic acid, degradation 446 447d
Nuclein 198
Nucleoid, in bacteria 489
Nucleolus 64 144
Nucleophile 232d
Nucleophilic attack 231 232d 249
Nucleophilic attack in DNA synthesis 465
Nucleoside, definition 201
Nucleoside, diphosphokinase 13t 348d
Nucleoside, diphosphokinase, location 13t
Nucleosome 217d 473
Nucleotidases 446
Nucleotide 202
Nucleus in adipocyte 164
Nucleus of actin 1324
Nucleus, l 2d 144 164 17 343
Numerical methods, in kinetic analysis 254
O-Glycoside 43s 44
Obligatory order, enzymatic reaction 259
Octadecanoyl-CoA 369s
Ogston, A, G. 359
Ogston’s three-point attachment proposal 3594
Oiled coil, elastin 126
Okasaki fragments 466 467 470
Oleate 369s 376\
Oleic acid 156s
Oleic acid, oleate 369s 376
Oleoyl-CoA 3694s
Oligomer 270s
Oligomeric protein 273
Oligomycin-sensitivity-conferring protein (OSCP) 412
Oligonucleotide 11t 25 206s 247
Oligonucleotide synthesis, solid phase 211d
Oligonucleotide, definition 44
Oligonucleotide, staining with PAS 21
OMP decarboxylase 438
Open system, definition 290
Operator, of gene 508
Opposite polarities of DNA strands 466
Optical activity 52
Optical activity of proteins 94 95
Optical activity, definition of 94
Optical activity, sugars 26 27
Optical rotation 44 52
Optical rotatory dispersion of proteins 94 95d
Optical rotatory dispersion, definition of 94
Optimum pH, of enzyme 261 262d
Order of reaction 274
Order of reaction, definition 252
Ordered BiBi, enzymatic reaction 2604
Ordered BiBi, reaction 280
Organic sulfate 11t
OriC 318d 460 463 470
Origin of the chromosome (oriC) 460
Origins of replication 472
Ornithine 70s 230s
Ornithine carbamoyltransferase 435
Ornithine carbamoyltransferase, location 13t
Orotate 437
Orotic aciduria 456
Osmium tetroxide 3
Osmotic lysis, of cells 19
Osmotic pressure 81 309d
Ostwald’s dilution principle 147
Outer membrane 13t
Outer membrane, mitochondrial 13t
Ovalbumin 102
Ovalbumin, molecular weight 83t
Overall reaction order 252
Ovomucoid, molecular weight 83t
Ovum 1
Oxaloacetate 324s 325s 374s 375s 422s
Oxaloacetate in gluconeogenesis 323s
Oxaloacetate in pyruvate carboxylase reaction 353s
Oxaloacetate in the citric acid cycle 346s 350s
Oxalosuccinate 347s
Oxidase, mixed-function 426
Oxidation of fatty acids 368
Oxidation reaction 295
Oxidation-reduction 230t
| Oxidative deamination 124
Oxidative phosphorylation, chemiosmotic model, Mitchell’s 408
Oxidative phosphorylation, definition 402
Oxidative phosphorylation, mechanistic models of 408
Oxidoreductase 230t 240
Oxolinic acid 475s
Oxonium ion 233s 233d
Oxygen 12 15
Oxygen 18, label 413
Oxygen affinity, hemoglobin 147 148
Oxygen in oxygenase reactions 12
Oxyhemoglobin 118
Palade, G. 4 13
Palade’s granules 13
Palmitate 369s
Palmitate, structure 153s
Palmitic acid synthesis 376 377t
Palmitic acid, 155s 156 376 396
Palmitic acid, palmitate 376
Palmitic acid, synthesis of 374
Palmitoyl thioesterase 377t
Palmitoyl-CoA 369ds 370d 371d 372t 395
Palmitoylcarnitine 370d
Pancreas 506
Pancreatic lipase 362 363d
Pancreatic peptidases 248
Pantothenic acid 56
Pantothenic acid in CoA 345s 346
Paracrystalline structures 18
Parental DNA 459
Parietal cell 427d
Partial reaction, linked to ATP 299d
Partial specific volume 81
Partitioning, of chromosome 458
PAS staining 21
PAS staining of erythrocytes 21
Pasteur effect 335d 418
Pasteur effect, connection to oxidative phosphorylation 418
Pasteur effect, effect on oxidative phosphorylation 412
Pasteur, L. 335
Payen 228
PCR, polymerase chain reaction 477—479 480 481d 487
Peas 419
Pectin 47
Pentamicrofibril 124d
Pentamicrofibril, tropocollagen 123
Pentose 25
Pentose phosphate pathway 16 241
Pentose phosphate pathway in erythrocytes 16
Pentose phosphate pathway, definition 339
Pepsin 229 248 426
Peptic cell 427d
Peptidase 11t
Peptide 53—75
Peptide bond 66 67 231 427
Peptide bond, structure of 61 d
Peptide hormones 427
Peptide, dipeptide 67
Peptide, enzymatic cleavage of 80
Peptide, glutathione 67d
Peptide, N-terminal 67
Peptide, naming of 67
Peptide, oligopeptide 67
Peptide, polypeptide 67
Peptide, tripeptide 67
Peptidyltransferase 504d
Perhydrocyclopentanophenanthrene 163s
Periodic acid Schiff (PAS) 2d
Periodic acid Schiff (PAS), staining 21
Peripheral protein, in membranes 172
Permease 174
Peroxidase 385d 452
Peroxisome 12
Persoz 228
pH effect, on enzyme rate 261d 262d
pH, definition of 57 58
Phagocytic vacuole 10d 22d
Phagocytic vacuole in macrophage 22d
Phagocytosis 21
Phagosome 10d 19
Phenanthrene 163s
Phenol 249
Phenomenase 241
Phenylacetate 455s
Phenylalanine 244 423t 425s
Phenylalanine, hydroxylase 425d
Phenylalanine, structure of 55t
Phenylisothiocyanate in protein sequencing 19d
Phenylisothiocyanate, Edman reagent 19s
Phenylketonuria 426 455
Phenyllactate 455s
Phenylpyruvate 426 455s
Phosphatase 11t
Phosphate 3sd 19
Phosphate, ester 2 233
Phosphate, esters of sugars 42
Phosphate, product of phosphatase reaction 19d
Phosphatide acid, phosphatidate 378
Phosphatide acid, synthesis of 380d
Phosphatidylcholine, cholesterol, acyltransferase (LCAT) 159t 365 379 380d 381d 382d 385d 397
Phosphatidylelhanolamine 159t 381d
Phosphatidylglycerol 159t
Phosphatidylinositol 159t
Phosphatidylserine 159r 381d
Phosphocreatine 456s
Phosphodiester linkage 204s
Phosphodiesterase 338 446
Phosphoelhanolamine 381d
Phosphoenolpyruvate in gluconeogenesis 325d
Phosphoenolpyruvate in glycolysis 316
Phosphoenolpyruvate, carboxykinase (PEPCK) 324 325d
Phosphoenzyme 232d
Phosphofructokinase 283 360
Phosphofructokinase in glycolysis 313
Phosphofructokinase, ADP, positive heterotropic effector 284
Phosphofructokinase, ATP, negative homotropic effector 284
Phosphofructokinase, citrate: negative heterotropic effector 284
Phosphofructokinase, control in gluconeogenesis 326d
Phosphofructokinase, control of 301d
Phosphofructokinase, control point in glycolysis 318 319d
Phosphofructokinase, thermodynamics of 300td
Phosphoglucomutase 327 329
Phosphoglucomutase in glycogen synthesis 327 360
Phosphoglucomutase, thermodynamics of 294
Phosphogluconate pathway 339
Phosphoglycerate dehydrogenase 425d
Phosphoglycerate kinase 315d
Phosphoglycerides, major classes 159t
Phosphoglycerides, structure 158s
Phosphoglyceromutase 315d
Phospholipase 362 363
Phospholipase 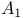 363 381 363 381
Phospholipase 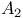 363d 381 385d 363d 381 385d
Phospholipase C 381
Phospholipase D 381
Phospholipid 11t
Phospholipid, absorption 362
Phospholipid, definition 158
Phospholipid, synthesis 379
Phosphomannose isomerase 330 331d
Phosphomevalonate 389d
Phosphomevalonate, kinase 389d
Phosphomonoester 11t
Phosphopantelheine 376
Phosphophosphorylase kinase 338d
Phosphoprotein phosphatase, in pyruvate dehydrogenase control 352 354d
Phosphoribosyltransferases 446
Phosphoryl-group transfer, thermodynamics of 298d
Phosphorylase a, from ‘b’ form 337 338d
Phosphorylase b, activation to ‘a’ form 337 338
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



 in
in