|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Kuchel P.W., Ralston G.B. — Schaum's outline of theory and problems of biochemistry |
|
|
 |
| Предметный указатель |
Phosphorylase b, thermodynamics of 295
Phosphorylase kinase 115 337d
Phosphorylase phosphatase 115
Phosphorylase, a, b 115
Phosphorylation 145
Phosphorylation in pyruvate dehydrogenase 116
Phosphorylserine 239
Phosphoserine phosphatase 425d
Phosphoserine transaminase 425d
Phosphosphingolipid 161s
Phosphotungstic acid, staining of tropocollagen 124d
Photoreactivation of DNA 476
Phylogenetic scheme 14
Piezoelectric effect 19
Ping pong, enzymatic reaction 259
Pitch of helix 107
Placenta, oxygen transfer across 147
Planck, M. 238
Plant 1
Plasma membrane 6d 7d 9d 16 19 21 129 134 150
Plasma membrane in adipocyte 16d
Plasma membrane, cilia 141 142d
Plasmalogen 159
Plasmid 217 461
Plasmodium 1
Plus end, of tubulin 140
Pointed ends, of actin filaments 139
Polar, helical rods 131
Polarimeter 26 52
Polarity, actin 133
polio virus 110
Poly( -D-glucosamine), chitosan 51 -D-glucosamine), chitosan 51
Poly(A) polymerase 497
Poly(A) tail on RNA 497
Poly-L-glutamate 103
Polyadenylation of RNA 497
Polyaminoacids 107
Polycistronic mRNA 494 512
Polydeoxynucleotide 206s
Polyhydroxy-aldehyde 25
Polyhydroxy-ketone 25
Polyisoprenoid 163
Polyketides, acetate as precursor 154
Polymer, of glucose 47
Polymerase chain reaction (PCR) 477—481d 487
Polynucleotide 204s
Polyproline 93
Polyproline helix 93
Polyribosome, polysome 505
Polysaccharidase 11t 25 44 46 126
Polysaccharidase as a substrate 11t
Polysome, polyribosome 13 505
Polyunsaturated fatty acid 155s
Pore 180d
Porin 174
Porphobilinogen 451s
Porphobilinogen, synthase 451
Porphyria 457
Porphyria, synthesis 451
Positive control of a gene 508
Positive cooperativity 269 273d 284
Positive cooperativity, enzyme kinetics and binding 266
Positive feedback control 284
Positive superceding of DNA 468
Positively cooperative binding 286
Posttranslational modification, of proteins 505 515 516
Potentia hydrogenii, pH 57
Potter — Elvehjem homogenizer 5
Pregnenolone 392s 393d
Prephenate 244s
Preproinsulin, structure 506d
Pribnow box 511 512 517
Pribnow box, DNA 492d
Primary active transport 179d
Primary lysosome 10d 20d 22d
Primary lysosome in macrophage 21d
Primary transcript 493
Primase (DnaG) 467 469d
Primer, DNA 465 479
Primidine nucleotide, binding to aspartate transcarbamoylase 115
Primosome 467
Principal of mass action 251 274
Procarboxypeptidase 427 428d
Prochirality 359
Procollagen, assembly 149
Procollagen, peptidases 123
Proelastase 248d 427
Progestogen 393d
Progress curve 252
Proinsulin, structure 506d
Proline 423t
Proline racemase 236
Proline racemase, planar and tetrahedral intermediates 236
Proline racemase, proline as substrate 236s
Proline, L-, D- 236s
Proline, structure of 55t
Prolylhydroxylase 505
Promoter 140 487 492
Promoter of gene 508
Promoter, binding 115
Promoter, mobile 467
Promoter, self association 112d
Proofreading of DNA 469
Prophase, lamins in 145
Propinquity, effect in enzymes 230 242
Propionate, structure 153s
Prostacyclin 384d
Prostaglandin 376 398
Prostaglandin,  384s 386d 384s 386d
Prostaglandin, 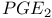 384s 386d 384s 386d
Prostaglandin,  153s 384s 386d 153s 384s 386d
Prostaglandin,  385ds 386d 385ds 386d
Prostaglandin, 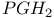 385d 386ds 385d 386ds
Prostaglandin,  384s 386d 384s 386d
Prostaglandin, structure 383
Prostaglandin, structures 384d
Prostaglandin, synthase 385
Prostanoic acid 383d
Prostate glands 4
Prosthetic group 81 232
Protease, specificity 11t 426 429t
Protein 2 76—107 108 117 228 386d
Protein 4 1 136 137d
Protein channel 7d
Protein conformation, of enzyme 239
Protein degradation 430d
Protein denaturants 87
Protein disulfide-isomerase 241
Protein evolution, phylogenetic tree 97 98d
Protein kinase (A) 337d
Protein structure 87—89
Protein structure,  - - - - motif 97d motif 97d
Protein structure, accessing via world wide web 99
Protein structure, determination of 99 100
Protein structure, domain structure 96
Protein structure, families of 96
Protein structure, folding domains 97
Protein structure, hierarchy of 95
Protein structure, planarity of peptide bond 88
Protein structure, primary 95
Protein structure, quaternary 96 97
Protein structure, Ramachandran plot 89d
Protein structure, secondary 95
Protein structure, stylized representation of 94d
Protein structure, supersecondary 95
Protein structure, tertiary 95
Protein structure, topology diagrams 96d
Protein synthesis 13
Protein synthesis, elongation 500 504d
Protein synthesis, enzymes 241
| Protein synthesis, inhibitors 500
Protein synthesis, initiation 500 504d
Protein synthesis, release factors, RF1, RF2 and RF3 505
Protein turnover 431
Protein,  -helix 112 -helix 112
Protein, affinity chromatography of 77
Protein, allosteric interactions 118
Protein, conformational change 115
Protein, conformational changes in 109
Protein, conformational entropy 84
Protein, connective tissues 108
Protein, cytoskeletal 108
Protein, denaturation with urea 81
Protein, dephosphorylation 115
Protein, dephosphorylation, in pyruvate dehydrogenase 116
Protein, dialysis of 77
Protein, disulfide bonding in 109
Protein, Edman degradation of 78 79
Protein, electrophoresis of 77
Protein, electrostatic interactions 84 85d 86t
Protein, folding 84 108
Protein, functions of 76
Protein, hydrogen bonds 85 86(
Protein, hydrogen bonds in 108
Protein, hydrophobic interactions 85 86t 108
Protein, ionic bonds in 108
Protein, packing contacts 117
Protein, phenylthiohydantoin derivative 79s
Protein, phosphorylation 115
Protein, phosphorylation, in pyruvate dehydrogenase 116
Protein, protein folding 108
Protein, purification of 76—78
Protein, quaternary structure 81 108 117
Protein, salt bridge 85d
Protein, salting out of 77
Protein, sequence homology 97 98
Protein, sequencing of 78—81
Protein, sliding contacts 117d
Protein, solubility of 77
Protein, structure dictated by sequence 87
Protein, supramolecular structure 108
Protein, van der Waals interactions 85 86t
Proteoglycan 121 126 127 128d 129
Proteoglycan, aggregated 129d
Proteoliposome 170 171
Proteolysis 145
Proteolysis in virus assembly 110
Proteolytic enzyme 148 229 426
Protofilament, of tubulin 139
Protomer 111
Protomer of binding protein 270
Proton motive force,  s 407 s 407
Proton motive force, definition 407d
Proton translocation in mitochondria by complex I 410
Proton translocation in mitochondria by complex III 410
Proton translocation in mitochondria by complex IV 410
Proton translocation in mitochondria, loop mechanism 410d
Proton translocation in mitochondria, not by complex II 410
Proton translocation in mitochondria, proton-motive Q cycle 410 411d
Proton-motive Q cycle 410
Protoporphyrin IX 451 452s
Proximal histidine, in hemoglobin 147
Proximity, effect in enzymes 230
Pseudo substrate 236
Pseudogene 499
Pseudomonas, bongkrekic acid from 415
Pseudouridine 218s
Purine 198s
Purine tautomer, enol 200s
Purine tautomer, imino 200
Purine tautomer, keto 200s
Purine, de novo synthesis 440d
Purine, inhibition of de novo synthesis 445d
Purine, metabolism 437
Purine, salvage synthesis 446d
Puromycin 507s 516
Pyran 35s
Pyranose 49
Pyranoside 44s
Pyridine 75s
Pyridoxal phosphate 232 421s 454
Pyridoxamine phosphate 421s
Pyrimidine 198s
Pyrimidine, bifunctional enzyme 438
Pyrimidine, biosynthesis by E, coli 229
Pyrimidine, CAD protein 438
Pyrimidine, de novo synthesis 438 439d
Pyrimidine, metabolism 437
Pyrimidine, pathway in ?, coli 438
Pyrimidine, salvage synthesis 446d
Pyrimidine, synthesis: control of 266
Pyrimidine, trifunctional protein 438
Pyrophosphatase 465
Pyrophosphatase in glycogen synthesis 328
Pyrophosphate 465
Pyrophosphomevalonate decarboxylase 389d
Pyrrole 2-carboxylate 236s
Pyruvate 240s 375d 424t
Pyruvate carboxylase 323d
Pyruvate carboxylase, allosteric activation by acetyl-CoA 353 354d
Pyruvate carboxylase, reaction 353
Pyruvate carboxylase, subunit association 309
Pyruvate decarboxylase 116 117 322 394 454
Pyruvate decarboxylase, 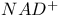 as substrate 116 as substrate 116
Pyruvate decarboxylase, (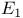 ) reaction 352 353d 3544 ) reaction 352 353d 3544
Pyruvate decarboxylase, acetyl-CoA as substrate 116
Pyruvate decarboxylase, CoA as substrate 116
Pyruvate decarboxylase, pyruvate as substrate 116
Pyruvate decarboxylase, subunit complex 116 117d
Pyruvate dehydrogenase complex 352—354
Pyruvate dehydrogenase complex, control of 302d 352
Pyruvate dehydrogenase complex, inactivation by phosphorylation 352 3544
Pyruvate dehydrogenase complex, mitochondrial location 352
Pyruvate dehydrogenase complex, overall reaction 352s
Pyruvate dehydrogenase kinase 352 354d
Pyruvate in glycolysis 311
Pyruvate in pyruvate carboxylase reaction 353s
Pyruvate kinase in glycolysis 3164
Pyruvate kinase, activation 318
Pyruvate kinase, control point in glycolysis 318
Pyruvate kinase, inhibition 318
Pyruvate kinase, positive feed forward control of 318
Pyruvate, complete oxidation of, thermodynamics 320d
Pyruvate, conversion to acetyl CoA 346
Pyruvate, conversion to elhanol 322sd
Pyruvate, conversion to lactate 321s 322d
Pyruvate, inhibition of membrane transport of 184
Pyruvate, metabolic fate of 319 320d
Q-structure 482
Quad, reaction 259
Quaternary structure 111 117
Quaternary structure, hemoglobin 147
Rabbit, cytochrome c 98fd
Racemase, active site 236
Racemic mixture 26 241
Radial spoke, in cilia 141 142d
Radioactive labels, study of citric acid cycle 358
Ramachandran plot 89d
Rapid-equilibrium reaction 281
Rapoport — Luebering shunt 344
Rat liver 4
Rate constant 237
Rate constant, definition 251
Rate enhancement in enzymatic reaction 233
Rate enhancement, enzyme 237 238 241 249
Rate of reaction 251
Rate-determining step 299
Reactancy, definition 259
Reaction coordinate 235d
Reaction order 274
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



 -D-glucosamine), chitosan
-D-glucosamine), chitosan 
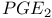


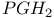

 -
- s
s 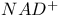 as substrate
as substrate 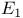 ) reaction
) reaction