|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Kuchel P.W., Ralston G.B. — Schaum's outline of theory and problems of biochemistry |
|
|
 |
| Предметный указатель |
Crossover theorem 300
Crossover theorem in studies of oxidative phosphorylation 418
Crowding, in solutions 114
CTP-choline: 1, 2-diacylglycerol, cholinephosphotransferase 380d
CTP: cholinephosphate, cytidylyltransferase 380d
Cyanide 229
Cyanide, antidote, nitrite 418
Cyanide, inhibition of electron transport chain 406d
Cyanide, poisoning, nitrite as antidote 418
Cyanogen bromide, reaction with proteins 80s
Cyclic AMP, cAMP in control of glycogenolysis and synthesis 336s 337d
Cyclic AMP, cAMP, structure 336s
Cyclohexane, boat 36s
Cyclohexane, chair 36s
Cycloheximide 501s
Cyclooxygenase 385d 398d
Cyclopentane 384 386
Cyclopropane fatty acid 182s
Cysteine 202s 233 423t
Cysteine, disulfide bond 68
Cysteine, modification by N-ethylmaleimide 69
Cysteine, oxidation of 68s
Cysteine, preventing oxidation of 69
Cysteine, reduction of 68s
Cysteine, S-carboxymethyl derivative of 69
Cysteine, structure of 55t
Cytidine deaminase 243sd 244
Cytidine deaminase, 5, 6-Dihydrouridine 243s
Cytidine deaminase, transition-state analog 244
Cytidine diphosphocholine (CDP-choline) 379d
Cytochemistry 3
Cytochrome 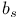 397 397
Cytochrome c, molecular weight 78 81
Cytochrome in electron transport chain 405t 406d
Cytochrome oxidase 405t
Cytochrome, 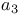 405t 406d 405t 406d
Cytochrome, a 405t 406d
Cytochrome, b 405t 406d
Cytochrome, c 404 405t 506d
Cytochrome, definition 404
Cytokeratins 145t
Cytokinesis 142
Cytokinesis, contractile ring in 144
Cytokinesis, plasma membrane in 144
Cytoplasm 7d 10d 19 20d
Cytoplasm, staining 19
Cytoplasmic membrane 21
Cytoplasmic membrane of macrophages 21 22d
Cytosine 12 198 199s 458
Cytoskeleton 6d 12 130 131
Cytoskeleton, actin in 137d
Cytoskeleton, anion channel attachment to 136 137d
Cytoskeleton, ankyrin 136 137d
Cytoskeleton, protein 4 1 136 137d
Cytoskeleton, proteins in 136
Cytoskeleton, spectrin in 136 137d 149 150
Cytoskeleton, submembrane 15
D loop in DNA 461
D-Alanine 240s
D-Amino acid oxidase 12
D-Erythro-pentulose 30
D-Erythrose 27 28 30
D-Erythrose, 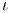 - 28 - 28
D-Erythrose, 4-phosphate 341s
D-Galactose 28s 47 49 51
D-Galactose, optical activity 52
D-Galactose, optical rotation 52
D-Gluconic acid 41s 47 51 52
D-Glucosamine, 2-amino-2-deoxy-D-glucose 42 51
D-Glucose 3 19 28s 29s 32s 33 47 123 396
D-Glucose in glycolysis 311
D-Glucose, anomeric carbon 33
D-Glucose, anomers  -, -,  - 32 34s 35s 48 - 32 34s 35s 48
D-Glucose, conformation 35 48
D-Glucose, Fischer projection 32s 34
D-Glucose, Haworth projection formulas 32 33 37t 48
D-Glucose, hexokinase and 237
D-Glucose, ionophore effects on transport 186
D-Glucose, NMR of 48
D-Glucose, open or straight chain structure 32
D-Glucose, optical rotation 48
D-Glucose, product of phosphatase reaction 19d
D-Glucose, reducing carbon 33
D-Glucose, transport across membranes 175 183 184
D-Glucose, transport modes 179t
D-Glucuronic acid 52
D-Mannitol 41s
D-Mannose 28s 49 51
D-Mannose in gluconeogenesis 330 331s
D-Ribitol 49s
D-Ribitol 5-phosphate 49 50s
D-Ribose 5-phosphate, in the pentose phosphate pathway 340s
D-Ribulose 30
D-Ribulose 5-phosphate 396
D-Ribulose 5-phosphate in the pentose phosphate pathway 340s
D-Threose 27 28
D-Xylulose 5-phosphate 240d
dAMP, deoxyadenylic acid 203
Dansyl chloride 19s
Daughter DNA 459
DCCD, dicyclohexyl carbodiimide 412
DCCD, dicyclohexyl carbodiimide, inhibitor of oxidative phosphorylation 412
ddATP 474
ddNTP 474
DEAE-cellulose 78s 102
Debye — Huckel theory 305
Defbrmylase 505
Degenerate code 489
Delft, in Holland 1
Denaturation of DNA 479 480d
Denaturation temperature, DNA 12
Density-gradient centrifugation 5
Deoxy sugars 39
Deoxyadenosine 221s
Deoxyadenylic acid, dAMP 203
Deoxycholic acid 168
Deoxycytidine 202
Deoxyguanosine 202s
Deoxyhemoglobin 119 121 148
Deoxyhemoglobin, cooperative effects 119
Deoxyhemoglobin, oxygen affinity 119
Deoxyhemoglobin, sigmoidal oxygen dissociation curve 119
Deoxyhemoglobin, tense structure 119
Deoxynucleoside triphosphate (dNTP) 465
Deoxyribonuclease (DNase) 219 446 447d
Deoxyribonucleoside 201
Deoxyribonucleotide 202
Deoxyribose 198
Deoxyribose 5'-phosphate 488s
Deoxyribose nucleic acid, DNA 198
Deoxythymidine 202
Deoxythymidine 5'-phosphate 202s
Depot fat, triacylglyerol in 158
Derivation of rate equation, algorithmic method 257
Derivation of rate equation, King and Altman 257
Derivation of rate equation, reaction patterns 257 258d
Desaturation, of fatty acid 376
Desmin 145t
Desmosine 126
Desmosome 145
Desolvation reaction 234
Detergent 363s
Detergent, properties of bile salts 168
Dextran 78
Dextrin 228
DFP (diisopropyl fluorophosphate) 239s
Diabetes mellitus 344
Diacylglycerol 158
Diad symmetry 111
Dialkyl phosphoamidite (DPA) 211
| Dialysis, in vesicle preparation 170
Diastase 228
Dicarboxylate carrier, in mitochondria 324
Dicarboxylate system, of amino acid transport 431
Dicyclohexyl carbodiimide (DCCD), inhibitor of oxidative phosphorylation 16ct 412
Dideoxy method of Sanger 473
dielectric constant 85
Dietary carbohydrate 374
Dietary lipid 391
Differential centrifugation 4 5d 24 303 308
Differentiation 1
Diffusion, coefficient 81
Diffusion, lateral, in membranes 173
Diffusion, rotational, in membranes 173
Digestion 228
Digestion of dietary lipid 391
Digestion of lipids 362 363
Digestion of triacyl glycerol 363sd
Digestion, role of bile salts in 391
Digestive enzyme, carboxypetidase A 247
Dihedral angle 111
Dihydrobiopterin 425d 426s
Dihydrobiopterin reductase 426d 455
Dihydrofolate reductase 443 444d
Dihydrofolate, dihydrofolic acid 448s
Dihydrolipoyl dehydrogenase (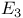 ), in pyruvate dehydrogenase 116 352 353d ), in pyruvate dehydrogenase 116 352 353d
Dihydrolipoyl transacetylase 352
Dihydrolipoyl transacylase (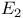 ), reaction 352 353d ), reaction 352 353d
Dihydroorotate 437
Dihydroorotate dehydrogenase 438
Dihydroorotate synthetase (or CAD) 438
Dihydroxyacetone 30 31
Dihydroxyacetone phosphate, in glycolysis 314s
Diisopropyl fluorophosphate (DFP) 239s
Dilution principle, Ostwald’s 147
Dimensional analysis 252
Dimerization of a protein 146
Dimerization, association constant 114
Dimethoxytrityl group (DMT) 211
Dimethylallyl pyrophosphate 389 390d
Dimethylallyl transferase 390d
Dinitrophenol, effect on oxidative, phosphorylation 412 416
Dipeptidase 430
Diphosphatidylglycerol 159t
Diphtheria toxin 507
Diploid 14
Diploid, cell 14
Disaccharide 25 44 126 127d 247
Discontinuous DNA replication 467
Disease, inherited 21
Dissociation equilibrium constant 255
Disulfide bond (S—S) rearrangease 241
Disulfide bond, bridge 80 240s
Dithiothreitol, reduction of disulfide bonds by 68s
DNA 7 11t 12 14 17 19
DNA as a substrate 11t
DNA gyrase 468 469d 470
DNA in nucleus 14
DNA ligase 467 468d 469d 476d 477 478d
DNA ligase, 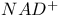 , reactant with 468 , reactant with 468
DNA mutation 470
DNA polymerase 465d
DNA polymerase  472 472
DNA polymerase 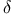 472 472
DNA polymerase III, holoenzyme 466 467 469d 470
DNA polymerase, I 465 469d 474d 476d 483
DNA polymerase, II 465
DNA polymerase, III 465 483
DNA primase 485
DNA synthesis 458
DNA, amplification 87 88d 464d
DNA, annealing 87 99 212 479
DNA, B form 209
DNA, base composition in various species 206t
DNA, base pairing 207
DNA, base-pairing geometry 207s 209
DNA, buckle 209
DNA, chain polarity 205
DNA, circular 459
DNA, cloning 238
DNA, complementary 207
DNA, COT analysis 214
DNA, denaturation 212d
DNA, denaturation temperature 12
DNA, deoxyribose in 198
DNA, deoxyribose nucleic acid 7
DNA, double helix 468
DNA, duplex 459
DNA, excision repair of 475
DNA, eye form 460
DNA, formamide effect on 212
DNA, helix 458
DNA, highly repetitive 498
DNA, hyperchromic effect 212
DNA, major groove 208 209d
DNA, melting temperature 214d
DNA, minor groove 208 209d
DNA, mitochondrial 12 461
DNA, moderately repetitive 498
DNA, mutation 458
DNA, negative supercoiling 468
DNA, nick in 464
DNA, nonhomologous recombination 463
DNA, nonrepetitive 498
DNA, Okasaki fragments 222
DNA, opening 209
DNA, photoreactivation of 476
DNA, pitch 209t
DNA, positive supercoiling of 468
DNA, primers 479
DNA, propeller 209
DNA, repair 458
DNA, repair synthesis 458
DNA, replication 458
DNA, replication fork 459
DNA, rise 208 209t
DNA, rolling-circle mechanism of replication 464d
DNA, satellites 15
DNA, semiconservative replication 459
DNA, separation by centrifugation 7
DNA, sequence heterogeneity 214
DNA, size 17
DNA, staining 2 19
DNA, structure 206d
DNA, template 481
DNA, termination of replication 460
DNA, thymine dimer in 475s
DNA, topology 215 459d
DNA, unstacking 212
DNA, uv damage 458
DNA, wild type 238
DNA, X-ray diffraction 208
DNA, Z-form 208 210d
DnaA 470
DnaA, boxes 470
DnaB 470
DnaB, helicase 468 469d
DnaC 470
DnaG 470
DNAhelicase 468 469d
Dnase, deoxyribonuclease 219
Dolichol 163s
Domain of protein 106
Dopa quinone 432s
Double helix, actin 113 132
Double-helical DNA 458
Double-reciprocal plot 253d 264d 266d
Down syndrome 15 24
Down syndrome, chromosomal translocations in 24
Down syndrome, facial features 15
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



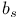
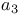
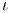 -
-  -,
-,  -
- 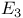 ), in pyruvate dehydrogenase
), in pyruvate dehydrogenase 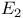 ), reaction
), reaction 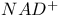 , reactant with
, reactant with