|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Kuchel P.W., Ralston G.B. — Schaum's outline of theory and problems of biochemistry |
|
|
 |
| Предметный указатель |
Fructose-1 -phosphate aldolase 331d
Fructose-1, 6-bisphosphatase 301d 314d
Fructose-1, 6-bisphosphatase in gluconeogenesis 325d
Fructose-1, 6-bisphosphate 242 314s
Fructose-1, 6-bisphosphate aldolase 242
Fructose-1, 6-bisphosphate aldolase in glycolysis 314d
Fructoside 43
Fumarase, fumarate hydratase 13f 249 348d
Fumarase, location 13f 436s
Fumarate, in the citric acid cycle 348s
Fumaric acid, oxidation energetics 304
Furan 35s
Furanose 49
Fusidic acid 507
G-actin 134d
GABA,  -amino butyrate 455 -amino butyrate 455
Galactitol 52
Galactokinase 330 331d
Galactosamine D-(2-amino-2-deoxy-D-galactose) 42
Galactose hydroxylysine 123
Galactose-1 -phosphate, uridylyltransferase 330
Galactoside 11t
Galactoside permease, in E. coli 508
Galactosylceramide 161s
Galactosylceramide- -galactosylhydrolase 383d -galactosylhydrolase 383d
Galactosyltransferase 149
Galacturonic acid 47
Gamete 14
Ganglioside 381 382 383
Ganglioside,  383d 383d
Ganglioside,  383d 383d
Ganglioside,  383d 383d
Ganglioside, definition 161
Gastric gland 427d
Gastrin 426
GATA-1, in erythroid cells 496
Gaucher disease 21 382f 383d
Gel electrophoresis, for determining, protein molecular weight 83
Gel filtration 102
Gel filtration of proteins 78 82 83d 83f
Gel filtration, molecular weight estimation 82f 83df
Gel-to-sol transition, actin 135
Gelatin 70 121
Gelsolin 135
Gene 458
Gene amplification 444 463
Gene cloning 477
Gene cluster 499d
Gene or DNA amplification 458
General acid catalysis 247
General acid-base catalysis 247
Generation-time of bacteria 462
Genes, isolation of 476
genetic code 489 490f 509
Genome 14 458
Geraniol 162s
Geranyl pyrophosphate 390d
Geranyl transferase 390d
Gibbs free energy 238 290 293
Gibbs free energy, change, AG 293
Gibbs, W. 238
Glial filaments 145t
Globin fold in hemoglobin 117 147
Globin, gene cluster 499d
Globin, synthesis 509 516
Globoside 162s
Glucagon, in glycogenosis 329
Glucan 45
Glucitol 51
Glucitol, D- 40s 41s
Glucocorticoid 393d
Glucofuranose,  -, -, - 35 - 35
Glucogenic amino acids 432
Glucokinase in liver 317
Glucokinase, 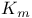 for glucose 317 318d for glucose 317 318d
Glucomannan 45
Gluconeogenesis, definition 323
Gluconeogenesis, scheme 324d
Glucose 1-phosphate, in glycogen synthesis 327
Glucose 6-phosphate 19 249t 317
Glucose 6-phosphate in glycogen synthesis 327
Glucose 6-phosphate in the pentose phosphate pathway 339
Glucose 6-phosphate, hexokinase and 237
Glucose-6-phosphatase 19 19t 249t
Glucose-6-phosphatase in gluconeogenesis 326d
Glucose-6-phosphatase, staining 19d
Glucose-6-phosphate isomerase, second step in glycolysis 31 3d
Glucoside 43
Glucuronate transferase 149
Glucuronic acid 126
Glutamate 420s 421s 422s 423t
Glutamate as pH buffer 72
Glutamate dehydrogenase 420 423d 434
Glutamate dehydrogenase, reaction 453d
Glutamate synthetase 420d
Glutamate, structure of 55t
Glutamic acid 233 247
Glutamic acid, isoelectric point 64t
Glutamic acid, pH titration of 63d
Glutamine 420s 421s 423t
Glutamine synthetase 420d
Glutamine synthetase, thermodynamics 299d
Glutamine, structure of 54t
Glutaraldehyde fixation 23
Glutathione 67s 240d 241
Glutathione synthetase 241
Glycan 45
Glyceraldehyde 30
Glyceraldehyde 3-phosphate 240 250
Glyceraldehyde 3-phosphate in glycolysis 314
Glyceraldehyde 3-phosphate in substrate level phosphorylation 402
Glyceraldehyde 3-phosphate in the pentose phosphate pathway 341
Glyceraldehyde kinase 331
Glyceraldehyde, D- 26 27
Glyceraldehyde, L- 26—28
Glyceraldehyde-3-phosphate dehydrogenase 241 315d
Glyceraldehyde-3-phosphate dehydrogenase and 1, 3-bisphosphoglycerate in glycolysis 315
Glyceraldehyde-3-phosphate dehydrogenase, active site 250
Glyceraldehyde-3-phosphate dehydrogenase, acyl compound 250
Glyceraldehyde-3-phosphate dehydrogenase, glyceraldehyde 3-phosphate in 250
Glycerol 363
Glycerol 3-phosphate 331
Glycerol 3-phosphate shuttle 333 334d
Glycerol in diphosphatidylglycerol 159t
Glycerol in gluconeogenesis 331 332
Glycerol in glycerolipids 157
Glycerol in phosphatidylglycerol 159t
Glycerol kinase 331 332d
Glycerol-3-phosphate dehydrogenase in glycerol 3-phosphate shuttle 333 334d
Glycerol-3-phosphate dehydrogenase, cytoplasmic 333 334d
Glycerol-3-phosphate dehydrogenase, mitochondrial 333 334d
Glycerolipids 154 157
Glycerose 29
Glycine 67s 238 240d 395 423t 424
Glycine in bile salts 168
Glycine, codon 149
Glycine, pH titration of 61d
Glycine, structure of 54t
Glycine, synthesis 424d
Glycocholate 168s 395
Glycogen 11t 17 23 46
Glycogen phosphorylase 329d
Glycogen synthase 328d
Glycogen, granules 327
Glycogen, site of synthesis 17
Glycogen, structure 328s 329d
Glycogen, synthesis of 327
Glycogenosis, control of 329 330d
Glycoglycerolipid, structure 160
Glycolipid 381
| Glycolysis in erythrocytes 15
Glycolysis, control points of 317
Glycolysis, definition 311
Glycolysis, effects of hormones on 336
Glycolysis, energetics of 316 317d
Glycolytic enzyme 242
Glycolytic flux 335
Glycolytic pathway 15
Glycoprotein 1 2
Glycosaminoglycan 47 122 126 127d 129d 130
Glycosaminoglycan, binding 131t
Glycosaminoglycan, chondroitin sulfate 127d
Glycoside 43
Glycosidic bond 25 42 45 50 245t
Glycosphingolipid 11 21 161 382d
Glycosyl bond 231t
Glycosylation 43 231
Glycosylation of collagen 123
Glycosyltransferase 126 149
Glycylalanine 67s 89
Glycylglycylglycine 75
Glyoxylate cycle, overall reaction 357d
Glyoxylate cycle, reactants 355 356d
Glyoxylate, in the glyoxylase cycle 356
Glyoxysome 356
Golgi apparatus 4 8d 9 9d 17 20d 22d 155s 156s 506
Golgi apparatus in macrophage 22d
Golgi apparatus, body 6d
Golgi apparatus, body size 17
Gomori, reaction 3 4
Gout 456
Gramicidin A 181
Granules, Palade’s 13
Granulocytes 130
Graphical evaluations, in enzyme kinetics 276
Graphical procedures, kinetic analysis 254
Group translocation, in transport 179d
Growth factor 129
GTP 140 504d
GTP, caps on tubulin 140
GTPtGDP, in the citric acid cycle 347d
Guanidine hydrochloride 87
Guanidinoacetate, synthesis 450d
Guanine 200s 458
Guanine, deaminase 447d
Guanosine 12 202
Gulose 28
Gut lumen 17
H, enthalpy 290 292
Hair 106 145
Haldane, J, B, S. 256
Half-cell, electrode potential 296 297d
Half-cell, reaction 295d
Halide bond 231t
Halobacterium halobium, membrane composition 172
Hanes and Woolf, plot 288 289
Haploid cell 14
Haworth projection formula, 3,6-anhydro-D-glucose 48 49
Haworth projection formula, D-glucose 32 33 37t 48
HDL, high-density lipoprotein 168 169t 390
Heart 17
heat capacity 292
Heat change, 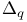 292 292
Helicase 476d
Helicase, DnaB 469d
Helix, A-form 209t
Helix, B-form 209t
Helix, buckle 2 10s
Helix, DNA 207 458
Helix, double 207
Helix, major groove 209d
Helix, minor groove 209d
Helix, opening 210s
Helix, pitch 107
Helix, propeller 210s
Helix, right-handed 207
Helix, roll 209 210s
Helix, tilt 209 210s
Helix, triple-stranded, collagen 121
Helix, twist 209 210s
Helix, weight tested 207
Helix, Z-form 209t
Helix-destabilizing protein, DNA 468
Hemagglutinin, trimer 112 113d
Heme 147
Heme in cytochromes 404
Heme, group 118d
Heme, synthases 451
Hemiacetal 32s 33s 43s
Hemicellulose 47d
Hemin 452 509
Hemoglobin 15 17 18 22 23 147 148 151 344
Hemoglobin tetramer 18
Hemoglobin, 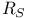 value 289 value 289
Hemoglobin,  chain 18 chain 18
Hemoglobin, 2, 3-bisphosphoglycerate binding 267
Hemoglobin, A, V, Hill and 267
Hemoglobin, Adair binding model 269
Hemoglobin, Adair equation and 289
Hemoglobin, allosteric interactions in 118 1
Hemoglobin, binding to 267
Hemoglobin, Bohr effect 120
Hemoglobin, BPG binding 147 151
Hemoglobin, comparison with myoglobin 151
Hemoglobin, cooperative effect 147 148
Hemoglobin, cyclic symmetry 11 8d
Hemoglobin, degradation 452
Hemoglobin, deoxyhemoglobin 148
Hemoglobin, dihedral symmetry 11 8d
Hemoglobin, electrostatic interactions 118
Hemoglobin, fetal 145
Hemoglobin, fractional saturation 267
Hemoglobin, globin fold 147
Hemoglobin, heterotropic effector 267
Hemoglobin, Hill coefficient 148 151
Hemoglobin, Hill equation and 289
Hemoglobin, hydrogen bonds 118 148
Hemoglobin, hydrophobic interactions 118
Hemoglobin, molecular weight 82t
Hemoglobin, MWC binding model and 271
Hemoglobin, oxy- 118
Hemoglobin, oxygen binding 119d 147 148 267
Hemoglobin, packing contacts 117d 118
Hemoglobin, proximal histidine 118 147
Hemoglobin, quaternary structure 97 118 147
Hemoglobin, relaxed structure 148
Hemoglobin, salt links 119
Hemoglobin, sedimentation coefficient 82t
Hemoglobin, sigmoidal binding curve 267
Hemoglobin, sliding contacts 117d 118 148
Hemoglobin, tense structure 121 148
Hemoglobin, x-ray analysis 97
Hemolymph, insect 52
Henderson — Hasselbalch equation 60 72 73
Henderson — Hasselbalch equation, derivation 306d
Heparan sulfate binding 131t
Heparin binding to fibronectin 130d
Hepatocyte 17
Hepatocyte, liver cell 12 17
Heptad repeat 150
Heptad repeat, spectrin 151
Hess’s law, of constant heat summation 304
Heterochromatin 509
Heterocyclic compound 2
Heterogeneous nuclear RNA (hnRNA) 497
Heterolytic bond cleavage 245 247
Heterophagy 9 10d 19
Heterotropic effect 267 271 273d
Heterotropic effector, definition 266
Hexadecanoate 369s
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



 -amino butyrate
-amino butyrate  -galactosylhydrolase
-galactosylhydrolase 


 -,
-,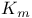 for glucose
for glucose 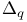
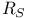 value
value