|
|
 |
| јвторизаци€ |
|
|
 |
| ѕоиск по указател€м |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Voit E. Ч Computational Analysis of Biochemical Systems: A Practical Guide for Biochemists and Molecular Biologists |
|
|
 |
| ѕредметный указатель |
"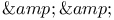 " declaration (of independent variable) 121 240 376 393 394 " declaration (of independent variable) 121 240 376 393 394
"at" (@) statement 110
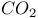 production 294 297 298 300 324 production 294 297 298 300 324
 228 228
 200 200
 21 43 64 142 145 159 160 165 169 188 374 418 444 21 43 64 142 145 159 160 165 169 188 374 418 444
 200 200
 200 200
 167 172 200 407 167 172 200 407
 27 262 263 264 275 283 288 366 27 262 263 264 275 283 288 366
 106 228 106 228
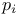 28 345 346 347 351 353 363 369 370 372 28 345 346 347 351 353 363 369 370 372
 178 178
 178 179 180 178 179 180
 -ketoglutarate 31 32 165 299 308 310 313 314 319 443 -ketoglutarate 31 32 165 299 308 310 313 314 319 443
 -ketoglutarate dehydrogenase 300 314 -ketoglutarate dehydrogenase 300 314
 -term 52 54 67 70 73 77 78 201 210 219 229 240 277 445 449 -term 52 54 67 70 73 77 78 201 210 219 229 240 277 445 449
 -5-phosphoribosyl amine 32 329Ч330 331 -5-phosphoribosyl amine 32 329Ч330 331
 -terms 23 50 51 52 70 73 77Ч80 82 108 194 201 210 219 229 237 240 244 247 265 267 287 446 450 462 463 -terms 23 50 51 52 70 73 77Ч80 82 108 194 201 210 219 229 237 240 244 247 265 267 287 446 450 462 463
 -terms aggregation 277Ч278 -terms aggregation 277Ч278
3-phosphoglycerate (3PG) 20Ч21 264
3-phosphohydroxyglycerate 23
3-Phosphohydroxypyruvate 20 22 23
5'(3')-nucleotidase (3NUC) 334 348
5'-nucleotidase (5NUC) 331 334 348
5-adenosylmethionine 32
Absolute change 22 55 68Ч69 71 225 230
Abstraction 57 326
Accumulation 46 64 77 109 128 132 133 167 262 295 345 402 408
Accuracy 58 59 67 83 92 106 107 148 149 157 158 226 315 319 336 340 357 388 395 398 404Ч410 419 444 450 461
Acerenza, L. 62 155 157 175 186 190 246 478
Acetate 249
Acetyl-CoA 188 297 308 309 310 311 313 314 320 457
Achs, M.J. 38 469 474
Aconitase 312 314
Activation and activators 54 55 115 126 135 183 215 327 444 see
Activation and activators allosteric 273 275 346
Activation and activators in anaerobic fermentation pathway 262
Activation and activators in maltose catabolic system 401
Activation and activators of carbamoyl phosphate synthetase 153
Activation and activators of degradation 220
Activation and activators of gene regulation 400 401 402 407
Activation and activators of pancreatic zymogen 29Ч31
Activation and activators of purine metabolism 334 349
Activation and activators of reaction 14 27 28 86 112Ч113
Activation and activators, branched pathway with 236
Activation and activators, feedback 198 206 220
Activation and activators, feedforward 279
Activation and activators, inhibition compared 4 220
Activation and activators, kinetic orders 145
Activation and activators, notation 14 27 55 349
Activation and activators, phosphorylase cascade 86
Activation and activators, strength of 216 380
Activation and activators, time-dependent 295
Ad hoc models 58
Ad hoc symbols 78
Adams, M.D. 8 473
Additive 3 219 406
Adenine (Ade) 56 146 247 331 333 335 336 338 340 342Ч345 347 351 352 363
Adenine (Ade) excretion 146 149Ч150 338 346 364
Adenine (Ade) oxidation 348
Adenine phosphoribosyltransferase (APRT) 328 329 334 341 348 349
Adenosine (Ado) 56 328 333 336 343 347 352
Adenosine deaminase (ADA) 331 334 348
Adenosine diphosphate (ADP) 27 56
Adenosine monophosphate (AMP) 56 90 128 263 271 272 274 275 290 328 330 331 333 335 336 346 347 349 351 352 454
Adenosine triphosphate (ATP) 12 14 27 28 56 90 128 150 152 153 167 168 187 188 247Ч249 262Ч264 267 269 270Ч272 274Ч278 283 290Ч292 297 327 328 330 333 336 342 343 346Ч348 352 362 366 454 458
Adenylate kinase 263
Adenylates 56 128 263 274 283 330 331 335 340 342Ч346 351 352 354Ч356 363 454 458
Adenylosuccinate (S-AMP) 56 330 331 333 336 338 346 347 352
Adenylosuccinate lyase (ASLI) 348
Adenylosuccinate synthetase (ASUC) 334 348
Adolph, E.F. 56 469
Adrenaline 6 38
Aerobic ATP formation 247
Affinity 109
Affymetrix 18
Aggregated kinetic order 85 87 267 338 339 342 355 357
Aggregation of fluxes 83 87 91Ч92 119 288
Aggregation of terms 265 267 272 351 448
Agresar, G. 129 469
Ahmad Mahir, B.R. 411 479
Akaoka, L. 359 474
Akerboom, T.P.M. 162 163 372 475
Akiyama, Y. 16 474
Alanine (ALA) 299 309 310 311 313 315 319 320 322 456
Alanine transaminase 300
Albe, K.R. 171 293 297 298 301 310 315 323 325 469 494
Alcohol 2
Alcon, C.H. 83 223 286 290 396 408 409 490
Algebra 62 167 173 191 200 209 252 391 395 406 407 413 414 426
Algebra Boolean 63
Algebraic analysis see "Algebraic steady-state evaluation"
Algebraic steady-state evaluation 61
Algebraic steady-state evaluation of glycolytic-glycogenolytic pathway in perfused rat liver 365Ч398
Algebraic steady-state evaluation of irregular S-systems 206Ч208
Algebraic steady-state evaluation of linear pathway with feedback 84 212Ч213
Algebraic steady-state evaluation of pathway with feedback and cross-activation 213Ч216
Algebraic steady-state evaluation of regular S-systems 83 119 121 201Ч206
Algebraic steady-state evaluation, branch point constraints 393Ч395
Algebraic steady-state evaluation, examples 212Ч216
Algebraic steady-state evaluation, exercises 217Ч221
Algebraic steady-state evaluation, kinetic order sensitivities 395
Algebraic steady-state evaluation, local stability 208Ч216
Algebraic steady-state evaluation, log gains of fluxes 385Ч388
Algebraic steady-state evaluation, log gains of metabolites 384Ч385
Algebraic steady-state evaluation, matrix notation 200
Algebraic steady-state evaluation, precursor-product relationships 391Ч393
Algebraic steady-state evaluation, rate constant sensitivities 390Ч395
Algebraic steady-state evaluation, steady-state equations 193Ч198 380Ч384
Algebraic steady-state evaluation, transition time 216Ч217 389Ч390
Algebraic steady-state evaluation, two-variable system 212
Algebraic steady-state evaluation, unconstrained parameters 391
Algebraic steady-state evaluation, zero steady states 198Ч200
Alginate-immobilized cells 260
Alignments, sequence 18
Alippi, C. 174 483
Allele 149
Allometry/allometric relationships 55Ч56 57 70Ч73 74
Allopurinol 353 360 361 364
Allosteric activator 273 275 346
Allosteric enzymes 163 271
Allosteric inhibition 76
Allosteric Michaelis-Menten reaction 302
Alphabet 62
Amendment of models 32 81 128 295 297 326 331 343 344 346 351 361
Amidophosphoribosyltransferase (ATASE) 329 334 340 348 361
AMP deaminase (AMPD) 334 340 345 348 355Ч356 357 363
Amphibolic pathways 25 92
Amplification/amplifiers 86 222 312 314 322
Amplitudes 110 111 116 117 118 121 125 137 409 449
Anaerobic fermentation pathway 260Ч261
Anaerobic fermentation pathway, activators 262
Anaerobic fermentation pathway, aggregation of  -terms 277Ч278 -terms 277Ч278
Anaerobic fermentation pathway, analysis of model 279Ч284
Anaerobic fermentation pathway, ATP dynamics, constraints 287Ч289
Anaerobic fermentation pathway, data 268
Anaerobic fermentation pathway, dynamics 282Ч283
Anaerobic fermentation pathway, enzyme concentrations 287Ч288
Anaerobic fermentation pathway, exercises 290Ч292
Anaerobic fermentation pathway, expansion of model 284
Anaerobic fermentation pathway, features 261Ч262
Anaerobic fermentation pathway, flux maximization 286Ч290
Anaerobic fermentation pathway, fructose-l,6-diphosphate degradation 273Ч275
Anaerobic fermentation pathway, glucose-6-phosphate degradation 271Ч273
Anaerobic fermentation pathway, glycogen synthetase reaction 272Ч273
Anaerobic fermentation pathway, GMA equations 265Ч268
Anaerobic fermentation pathway, hexokinase step 269Ч271
Anaerobic fermentation pathway, linear programming 285Ч286
Anaerobic fermentation pathway, logarithmic gains 280Ч282
| Anaerobic fermentation pathway, mass conservation 288Ч289
Anaerobic fermentation pathway, metabolite concentration 288
Anaerobic fermentation pathway, objective function 286Ч287
Anaerobic fermentation pathway, optimization 284Ч286 289Ч290
Anaerobic fermentation pathway, parameter values 268Ч277
Anaerobic fermentation pathway, phosphofructokinase reaction 271Ч272
Anaerobic fermentation pathway, properties of model 279Ч284
Anaerobic fermentation pathway, pyruvate kinase reaction 275
Anaerobic fermentation pathway, robustness of model 283Ч284
Anaerobic fermentation pathway, S-system representation 277Ч278
Anaerobic fermentation pathway, sensitivities 282
Anaerobic fermentation pathway, steady state 280 287
Anaerobic fermentation pathway, sugar transport 268Ч269
Anaerobic fermentation pathway, variables defined 262Ч265
Analytical evaluation 14 83
Analytical solution 65Ч66 75 210 350 353 406 410
Anderson, M.L. 315 325 493
Anderson, S.P. 17 472
Animal experiments 8 362 365 401
Antagonistic interactions 11
Anton, H.J. 410 491
Apparent first-order rate constant 308
Appel, R.D. 9 493
Appenzeller, T. 5 474
Approximate character 8 3
Approximation in biochemical system models 37Ч38 57 88 90 142 144 147 160 164 184 225 226 290 338 357 360 403 404 409 414 417Ч421 425 426 440 447 448 450 466 467
Approximation of degradation 59
Approximation second-order 406
Approximation theory 56 57
Apweiler, R. 18 469
Arcs 64
Arginine 401
Argonne National Laboratory 16
Aromatic amino acid 20
Arrayer 17
arrows 151 315
Arrows arched 135
Arrows bi-directional 28Ч29
Arrows curved 349
Arrows dashed 443
Arrows dotted 443
Arrows double-headed 25 26 27 28Ч29 346
Arrows double-tailed 27 28Ч29 87 346
Arrows, activation 349
Arrows, feedback control 4 13 251
Arrows, flow 14 20 23 25 27 28Ч29 31 93 130 133 251 443
Arrows, flux 25 26 27
Arrows, improper diagrams 14 31 443
Arrows, inhibition 349
Arrows, modulation 26 27 31 155
Arrows, signal 443
artificial intelligence 61
Aspartate  -semialdehyde 32 -semialdehyde 32
Aspartate (ASP) 32 299 300 309 310 315Ч317 319 325 457 458
Aspartate transaminase 300 308
Aspartyl phosphate 32
Aspergillus niger 162 167Ч171
Associative 3 23 414 439
assumptions 37 38 39 49 58 72 128 139 144 171 178 241 283 307 322 325 326 330 343 363 379 391 396 406
Assumptions, quasi-steady-state 37
Asymptomatic 364
ATP turnover 187 247 248 249
Auscher, C. 364 472
Australian National Genomic Information Service (ANGIS) 18
Autogenous circuits 400
Automata theory 63
Auxiliary variables 102 138 220 370
Averet, N. 323 483
Ayvazian, J.H. 343 345 469
Backward reaction 98
bacteria 8 11 17 295 327 405
Bailey, J.E. 61 187 260 261 268 269 270 273 396 405 409 474 475 476 487
Bairoch, A. 15 18 469
Baker's yeast 8
balance 52 72 95 195 196 283 296 317 323 333 334 353 370 438
Balance equation 61 93 94 267 371
Balthis, W.L. 140 411 469 491
Bangerter, J.K. 140 469
Barber, J.R. 343 470
Barker, J.L. 63 493
Barker, W.C. 18 470 474
Barrell, B.G. 8 9 475
Barry, C.E., 3rd. 8 471
Bartel, T. 327 470
Bartelmess, I.B. 149 473
Baseline 109 112 114 116 140 141 142 148 228 232
Baseline experiment 52 115
Basmanova, S. 15 487
Bassham, J.A. 367 373 374 470
batch processes 22 23 412
Batschelet, E. 56 470
Battey, J. 8 470
Baxevanis, A.D. 15 470
Becker, M.A. 355 359 364 470 477
Bel-Enguix, G. 62 471
Benson, D.A. 18 470
Berg, P.H. 174 183 470
Bernstein, EC. 18 470
Berry, M.N. 415 421 422 470
Best fit 151 174
Best-case scenario 140
Bhalla, U.S. 3 171 493
Bi bi 301 302
Bi-directional arrows 28Ч29
Bi-directional processes 315 325
Bi-phasic response 107
Bi-substrate reaction 46 47
Bicyclic glutamine synthase cascade 86
Bimolecular reactions 53 87 88Ч89 171
Bin Razali, A.M. 175 176 189 404 490
Binns, M.R. 297 484
Biochemical interpretation 8 9
Biochemical map see "Maps biochemical"
Biochemical parameters 63 see
Biochemical reactions 55
Biochemical signals 9 14 17 47 53 63 86 93 187 248 284 314 320 330Ч331 397 444
Biochemical system models 7
Biochemical system models, alternatives to S-system models 58Ч64
Biochemical system models, analytical convenience 58
Biochemical system models, approximations 37Ч38
Biochemical system models, change in dependent variables 39 41Ч49 51 56
Biochemical system models, computer simulations 38Ч39
Biochemical system models, differential equations 42 43 45 47 48 65Ч67
Biochemical system models, examples 52 54Ч55
Biochemical system models, exercises 64Ч65 75
Biochemical system models, integrated 39 40Ч41
Biochemical system models, kinetic orders 37 53Ч54 55
Biochemical system models, linear relationships 56
Biochemical system models, mechanistic approach 39
Biochemical system models, networks as languages 62Ч64
Biochemical system models, parameters 39 51Ч55
Biochemical system models, power-function representations 59Ч60
Biochemical system models, rate constants 51Ч52
Biochemical system models, rate laws in 37Ч43 47
Biochemical system models, relative changes in flux 73Ч75
Biochemical system models, S-system form 49Ч51 55Ч58 65Ч75
Biochemical system models, stoichiometric network form 60Ч62
Biochemical system models, theoretical justification 57Ч58 60 70Ч73
Biochemical system models, validity 55Ч57
Biochemical system models, variables 17 24 25 38 39 48 56
Biochemical time scales 6 209 307 403 404
Bioinformatics 8 9 16 18 19
Biomass allocation 404
Biomolecular structures 18
Biomolecular time scales 6
Biotechnological optimization 83 260 285 390 397
biotechnology 18 22 62 64 223 284 287 290 400
Bish, D.R. 171 470
Bittner, M. 18 473
Blecker, J. 410 491
|
|
 |
| –еклама |
 |
|
|
 |
|
ќ проекте
|
|
ќ проекте



 " declaration (of independent variable)
" declaration (of independent variable) 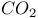 production
production 
 -ketoglutarate
-ketoglutarate  -5-phosphoribosyl amine
-5-phosphoribosyl amine