|
|
 |
| јвторизаци€ |
|
|
 |
| ѕоиск по указател€м |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Voit E. Ч Computational Analysis of Biochemical Systems: A Practical Guide for Biochemists and Molecular Biologists |
|
|
 |
| ѕредметный указатель |
O-succinylhomoserine 32
Objective function 174 286Ч287
Oden, K.L. 342 482
Ogata, H. 15 16 62 475 482
Ogiwara, A. 16 474
Oil palm 174
Okamoto, M. 174 223 400 482 489
Oleate 151 152
Oligomeric 135
Olivier, J.-L. 364 472
Olsen, L.E 57 482
Oosterhof, A. 345 477
Open systems 93Ч94
Operating point 60 69 83 84 88 91 96 137 140 159 160Ч162 164 165 186 189 248 269 288 297 341 355 356 361 371 408 409 417Ч420 425 426 440 447 456 466 467
Operations research 62 284
Operon 62
Optimal solution 285 289 409
Optimality, criterion of 285 389
Optimization 59 62 261 288 396 408Ч409
Optimization, anaerobic fermentation pathway model 284Ч286 289Ч290
Optimization, biotechnological 83 260 285 390 397
Optimization, linear programming 285Ч286
Orbit 111 114
Orcutt, B.C. 18 470
Order-of-magnitude model 145Ч146
Order-of-magnitude values 149 160 311 335
ORGANIZATION 2 5 7 11 98 129 402
Organizational level 6 11
Organized complexity 4 5
Ornithine 32 153 154 155
Ornithine transcarbamoylase 153 154 155
Ornithine-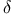 -aminotransferase 32 -aminotransferase 32
oscillations 4 45 51 57 61 63 64 110Ч111 113 117 118 124Ч125 216 224 279 283 307 376 409 410 448 449
Oscillations, damped 114 116 448
Oscillatory behavior 209 216 280 340
Ostell, J. 18 470
Ou, K. 9 493
Ouellette, B.F.F. 15 18 470
Output 38 45 61 63 64 93 103 105 106 111 114 122 129 134 139 140 193 216 232 280 290 326 390 396 408 412
Outside the system 22 93 94
Ovadi, J. 171 482
Overbeek, R. 15 16 487
Overdetermined 190
Overexpression 290 327
overproduction 340 345
Overshoot 84 308 378 417
Oxalacetate (OAA) 151 152 153 188 298 299 300 308 309 323 324 325 450
Oxidative pentose pathway 262
Oxidized form 27
Oxipurinol 360 361
Oxygen consumption 294
Oxygen uptake 248
Oxypurines 56 364
Page, T. 343 482
Paley, S. 16 478
Palsson, B.0. 61 62 129 209 284 482 486 490
Palsson, H. 129 209 482
Pancreas 24 29Ч31
Pandolfi, P.P. 17 482
Panetta, J.C. 141 482
Panyushkina, E. 15 16 487
Paper and pencil computation 65 97 105 119 124 143 280 406
Papoutsakis, E.T. 284 482
Paradigm 3 37 417
Paradigm shift 4 5Ч6
Parameter estimation 60 143Ч145 175 328 333 335 340 351 365 369 396 405 427
Parameter estimation by double modulation 157Ч158
Parameter estimation for branched pathway 176Ч179
Parameter estimation for cascades 179Ч184
Parameter estimation for in vivo systems 171Ч172
Parameter estimation from dynamic data 172Ч185
Parameter estimation from steady-state data 146Ч158 191Ч192 335Ч338 368Ч370
Parameter estimation from traditional rate laws 158Ч164
Parameter estimation, adenine excretion 149Ч150
Parameter estimation, citrulline pathway 152Ч155
Parameter estimation, constraints used for 191Ч192 370Ч372
Parameter estimation, examples 149Ч155 165Ч171 191Ч192
Parameter estimation, exercises 185Ч189
Parameter estimation, experimental measurements 146Ч149 155Ч158
Parameter estimation, flux stoichiometry 191
Parameter estimation, genetic algorithms 174
Parameter estimation, glucose-6-phosphate in Aspergillus niger 167Ч171
Parameter estimation, isocitrate in Dictyostelium discoideum 165Ч167
Parameter estimation, kinetic orders 53Ч54 55 146Ч164 190Ч191 338 372Ч375
Parameter estimation, logarithmic gains 121 155Ч158
Parameter estimation, purine metabolism 335Ч338
Parameter estimation, pyruvate carboxylate in rat liver 151Ч152
Parameter estimation, pyruvate kinase in rat liver 150Ч151
Parameter estimation, rate constants 164Ч171 375
Parameter estimation, regression methods 173Ч174
Parameter estimation, S-system equations 375
Parameter estimation, scope of expected results 145Ч146
Parameter estimation, slopes replacing derivatives 174Ч184
Parameter estimation, transient responses 184Ч185
Parameter values 21 27 39 52 58 60 76 79 82 85 92 95 96 115 116 121 130 135 136 138Ч140 146 157 158 168 172Ч176 179 182Ч184 186 187Ч189 191 194 206 212 220 222Ч224 234 236 259 279 280 282 283 291 307 316 317 319 322 330 335 345 346 362 363 377 383 391 393 396 403 405 407 421 431 445 447 449 451 457
Parameter values and dependent variables 122Ч123 143 144
Parameter values in computer simulations 98 99 101
Parameter values, anaerobic fermentation pathway 268Ч277
Parameter values, changes in 109Ч110 122Ч123 379Ч380
Parameter values, glucose-6-phosphate metabolism 99
Parameter values, glycolytic-glycogenolytic pathway 368Ч375 379Ч380
Parameter values, inhibition 113 125
Parameter values, tricarboxylic acid cycle 298Ч306
Parameterize 174 341
Parameters 4 40 65 97 103 105 107 112 125 126 127 159 193 197 210 213 233 235 239 257 272 311 314 343 344 353 395 402 404 464 466
Parameters, constrained 71 87 191Ч192 241 243 245 287Ч288
Parameters, defined 22
Parameters, graphical representation 21 22 23
Parameters, rate law 2 45
Parameters, S-systems 50 51Ч55 58 60
Parameters, sensitivity 122Ч123 222 244Ч245
Parameters, unconstrained 390
Park, D J.M. 296 494
Parkhill, J. 8 471
Partial derivatives 88 89 166 167 170 225 230 241 304 318 394 414 424 425 440 460 461
Partial differentiation 89 91 147 161 165 169 268 275 277 304 371 446 447 451 456 467
Particle Physics 6
Pasquali, C. 9 493
Passonneau, J.V. 159 480
Path computation tool 16
PathComp 16
Pathological condition 40
Pathways 47 see "Branched "Linear "Superpathways"
Pathways amphibolic 25 92
Pathways converging 77 87 95
Pathways didactic 115 127 176 256 405
Pathways diverging 95 267 288
Pathways metabolic 15Ч16
Pathways, citrulline 152Ч155
Pathways, clamped 92 96
Pathways, graphical representations 15
Pathways, physiological shortening of 57
Pathways, polyamine 342
Pathways, reversible 91Ч93
Pathways, surprise 115Ч119
Patnaik, L.M. 174 488
Patrinos, A. 8 9 472
Payne, W.E. 16 477
Pelligrini-Toole, A. 16 478
Pentose 262 284
Pepsin 160
Permanent change 222 233 462
Permeabilized cells 148
Persistent changes 108Ч109 117Ч118 155 222 231 233 378
Perturbations 17 39 103 114 117 118 125 130 143Ч145 155Ч157 172 184Ч186 208 209 222 223 231 279Ч281 283 296 297 306Ч308 310 312 319 321 325 328 340 344 345 354Ч355 357 395 397 398 402 403 405 406 408 457 458 464 see
Perturbations and local stability 124
Perturbations in dependent variable 377Ч378
Perturbations in independent variable 109 378Ч379
Perturbations, log gains/sensitivities and 123
Peschel, M. 56 60 129 410 482
| Peterson, J.L. 64 482
Petkov, S.B. 409 482
Petri nets 63Ч64
Pfeiffer, E 18 470 474 481
Ph 23 24 27 148 260 270 271 272 275 281 283 286 292 415
Pharmacogenomics 9
Phase plane 110Ч111 114 116 118 119 126 250 409 448
Phase plot 250
Phillips, J.W. 415 421 422 470
Phosphatase 21 22 23 25
Phosphate (P,) 12 100 153 154 155 284 346 347 353 369 385
Phosphoenolpyruvate (PEP) 90 150 151 152 262 264 273 274 275 280 283
Phosphoenolpyruvate carboxykinase (PEPCK) 152
Phosphoenylpyruvate (PEP) 150
Phosphofructokinase (PFK) 28 29 100 101 109 122 125 262 264 267 271Ч272 278 281 282 283 291 367 369 370 372 374 376 390 391 398 447 455 456
Phosphoglucomutase 28 29 100 122 366 367 370 372 384 390
Phosphoglucose isomerase 28 29 100 367 370 374 377 391 398
Phosphoglycerate dehydrogenase 20 23
Phosphohomoserine 23 27 32
Phosphoribosyl pyrophosphate synthetase (PRPPS) 329 334 348 355 357 361 364
Phosphoribosyl pyrophosphate synthetase superactivity 340 355 357
Phosphoribosyltransferase 328 329 334 341 348 349
Phosphorylase 28 29 86 295 331 367 368 370 372 384 396 398
Phosphorylase a 100
Phosphorylation 63 64 148 295 379 380
Phosphorylization 24 249
Phosphoserine 20 21 22 23
Phosphoserine phosphatase 20Ч21 22 23
Phosphoserine transaminase 20 23
Physiological variation 57 356
Physiology 1 86 458
Physiomics 9
Pickard, W.E 56 483
Ping pong 301 302 303
Pinkhas, H. 345 477
Places 64
PLAS program see "Computer simulations"
Plasmids 17 149
Plesser, T. 272 275 476
plot 43 80 108 110 111 114 116 117 118 125 126 146 147 149 175 187 311 423 448 449 450 451
Pogson, C.I. 149 488
Polyamine pathway 331 342 343
Polymerase 348 351 363
Polymerase chain reaction 17 363
Polymorphisms 9 17
Polynomials 60 67 69 210 413 418 419 439 440 466
Polynomials characteristic 211 212 213 453
Polysaccharides 262 263 264 267 271 272 276 278 282 285 291 455 456
Pools/pooling 14 20 21 23Ч25 27 45 46 50 56 79 83 91 83 90 119 132 135 136 144 206 221 230 263 290 297 298 299 309 314 316 322 324 328 330 331 333 335 337 340 342Ч344 346 353 354 357 361 363 403 434 444 447 449 458 see
Pools/pooling combined 128 296
Pools/pooling, condensation of 129Ч131
Pools/pooling, sizes 42 49 51 296 315
Population model 7
Porteous, D.J. 149 473
Potter, C. 337 364 490
Power-function representation 41 50 53Ч54 57 59 88
Power-law approximation 90 144 160 164 338 414 419Ч421 425Ч426 440 447 467
Power-law equations 50
Power-law functions 49Ч50 55 60 74 77 102 138 142 155 164 176 192 198 251 303 305 368 389 390 398 404 406 407 408 413 420 425 426 448 452 460 467
Power-law functions, products of 56Ч57 59 70 73
Power-law rate law 167 168 451
Power-law terms 73 77 80 86 91 95 103 137 159 160 164 165 171 177 188 269 271 277 291 334 338 347 370 375 394 444 455 465 466
Powers 50
PP-ribose-P 12
PP-ribose-P synthetase 12
Precursor 67 78 79 86 87 115 119 208 285
Precursor-product constraints 79Ч80 82 83 99
Precursor-product relationships 79 84 90 99 111 115 130 132 176 184 190 193 240 266 267 271 290 322 371 391Ч393
Prediction 64 65 98 117 122 126 128 140 152 226 230 234 236 243 251 262 290 295 296 297 307 323 324 325 326 341 353 356 377 384 387 390 392 399 401 402
Preissler, H. 40 474
Prestalk cell 293
Pring, M. 38 474
Procarboxypeptidase 29Ч30
Product notation 51
Production 25 26 31 47Ч49 57 59 68 72 77 78 81 86Ч109 119 126 149 153Ч155 191 193 208 236 237 247Ч249 260 262 264 267 271Ч273 276 278 281 282 284Ч286 288Ч291 294 295 304 308 316 329 344 351 360 363 366 391 408 415 421 422 448 450 451 455 456 457 461 465
Production function 51
Production parameters 50 53 55
Production term 50 51 54 55 70 94 133 265
Productivity 128 131Ч133 134 141 389 396
Proelastase 30 31
Profile, expression 9 18
Prokaryotic Database 18
Pronevitch, L. 15 16 487
Proofreading 223 400
Protein 7 9 11 15 19 63 152 168 297 298 300 304 324 325 348 369 400 457 458
Protein catabolism 294 315 317
Protein Data Bank (PDB) 18
Protein degradation 294 314 315Ч316
Protein in serum 40
Protein Information Resource (PIR) 18
Protein O-methyltransferase (MT) 348
Protein sequences 16 18
Proteomes 9
Proteomics 9 16
PRPP synthetase superactivity 340
Pseudo-three-dimensional representation 18 123 228 237 243 249 311 320 324 325 423
Pseudoplasmodium 293
Puig, J.G. 355 470 477
Puigjaner, J. 60 246 383 483
Purine biosynthesis 13 328
Purine metabolism 12 56 149 223 326Ч328 364 399 405
Purine metabolism, activation of 334 349
Purine metabolism, adenylate turnover 343
Purine metabolism, AMPD inhibition 355Ч356
Purine metabolism, biochemistry 328
Purine metabolism, computer simulations 340Ч341 353Ч361
Purine metabolism, degradation 330
Purine metabolism, drug treatment simulation 360Ч361
Purine metabolism, enzyme activity changes 355Ч360
Purine metabolism, equation computation 338Ч340
Purine metabolism, equation construction 332Ч338
Purine metabolism, exercises 362Ч364
Purine metabolism, HGPRT deficiency 356Ч360
Purine metabolism, input changes 353Ч354
Purine metabolism, kinetic characteristics 338
Purine metabolism, models 328Ч361
Purine metabolism, modifications to model 342Ч343
Purine metabolism, parameter estimation 335Ч338
Purine metabolism, polyamine pathway 342
Purine metabolism, processes 332Ч335 346Ч353
Purine metabolism, PRPP synthetase superactivity 355
Purine metabolism, sensitivity analysis 342
Purine metabolism, steady-state concentrations 335Ч337
Purine metabolism, steady-state fluxes 337Ч338
Purine metabolism, temporary perturbations 354Ч355
Purine nucleoside phosphorylase 331
Purine ring 330 331 349
Purine synthesis 12 328 329 334 344 359
Purine synthesis de novo 12 329
Pyridoxal phosphate 32
Pyrimidine 344 350 362
Pyrophosphate 12 328 329
Pyrroline-5-carboxylate 32
Pyruvate (Pyr) 22 90 150 151 153 249 262 273Ч276 280 283 287 288 297 300 308Ч310 313Ч315 320 322Ч324 456 457 461
Pyruvate carboxylase 150 151Ч152 153
Pyruvate dehydrogenase 300
Pyruvate kinase 150Ч151 262 273 274 275 276 280 283 287
Qualitative behavior 4 283
Qualitatively correct 341 362
Qualitatively different 74 113 118
Quantitative laws 7 20
Quantitative terminology 19Ч20
Quasi-steady-state assumption 37
Quick, W.P. 149 489
Rabitz, H.K. 222 483
Radioactive decay 3
Ramos, T.H. 355 477
Random number 411
Randomness 3
Rank correlation 140 247Ч249
|
|
 |
| –еклама |
 |
|
|
 |
|
ќ проекте
|
|
ќ проекте



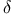 -aminotransferase
-aminotransferase