|
|
 |
| јвторизаци€ |
|
|
 |
| ѕоиск по указател€м |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Jacob C. Ч Illustrating Evolutionary Computation with Mathematica |
|
|
 |
| ѕредметный указатель |
RNA chromosomes, silent mutations 117
RNA chromosomes, summary of encoding by RNA triplets 90
RNA chromosomes, translation of codons to proteins 88Ч90
RNA chromosomes, triplet encoding of amino acids 88
RNA chromosomes, triplet table over RNA alphabet 88Ч89
RNA chromosomes, visualization 91Ч92
RNA chromosomes, with nucleotide bases 91Ч92 94
Roll turtle move operations 461
Rozenberg, G. 439 486
Rules,  reduction rules 404 406 reduction rules 404 406
Rules, 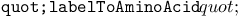 rules 89 90 rules 89 90
Rules, 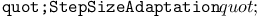 rule 226 227 rule 226 227
Rules, 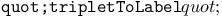 rules 89 rules 89
Rules, ArtFlower L-rules 495Ч497 498 509
Rules, cellular automata 441 442 446
Rules, classifier rule 289
Rules, DOL-systems 450 451 452 454
Rules, hierarchical GP-rule systems 292
Rules, Hilbert rules 463
Rules, IL-systems 458 473
Schema theorem for GA 190Ч207
Schema theorem for GA, as fundamental theorem of GA 205
Schema theorem for GA, covariance and selection theorem vs 207
Schema theorem for GA, defining length of a schema 194
Schema theorem for GA, described 190
Schema theorem for GA, enumeration of all schema instances 179 199Ч202
Schema theorem for GA, experiments with schemata 194Ч202
Schema theorem for GA, extraction of schema instances 197
Schema theorem for GA, GA-deceptive problems 206
Schema theorem for GA, generating schema instances for a population 197
Schema theorem for GA, generating schema instances over an alphabet 196Ч197
Schema theorem for GA, generating schemata of fixed length 194Ч196
Schema theorem for GA, instances of a schema 193
Schema theorem for GA, inversion and 206
Schema theorem for GA, mutation resistance 205
Schema theorem for GA, order of a schema 194
Schema theorem for GA, overview 205Ч206
Schema theorem for GA, problems with 206Ч207
Schema theorem for GA, recombination survival 204Ч205
Schema theorem for GA, schema defined 193
Schema theorem for GA, schema fitness 203Ч204
Schema theorem for GA, schemata as building block filters 202
Schema theorem for GA, schemata defined 190
Schema theorem for GA, selection filter survival 203Ч204
Schema theorem for GA, survival of building blocks 202Ч206 575
Schemata see schema theorem for GA
Schwefel, Hans Ч Paul 212 225 279
Search space for biomorphs 47
Second-order evolution 226 229
Selection (see also evolution strategies)
Selection and reproduction schemes selection,  selection templates 407Ч408 selection templates 407Ч408
Selection functions 161Ч167
Selection functions,  function 166Ч167 function 166Ч167
Selection functions,  function 163Ч164 function 163Ч164
Selection functions,  function 165Ч166 function 165Ч166
Selection functions,  function 323Ч325 function 323Ч325
Selection functions, additional functions 167
Selection functions, elitist selection 166Ч167
Selection functions, fitness-proportionate selection 162Ч164
Selection functions, plus and comma strategies 162 171 190 191Ч192
Selection functions, random survival 162
Selection functions, rank-based selection 164Ч166
Selection functions, tournament selection (FSA) 323Ч325
Selection, as principle of evolution 2 6
Selection, cumulative 5Ч33
Selection, directional 34
Selection, elitist 166Ч167
Selection, EP selection and evolution scheme 322Ч325
Selection, ES graphical notation for 253
Selection, ES selection and reproduction schemes 252Ч256
Selection, evaluation and 59
Selection, fitness-proportionate 162Ч164
Selection, GA evolution schemes and 161Ч169
Selection, GA selection 168Ч169
Selection, GP selection 372Ч373
Selection, in evolutionary parameter optimization scheme 73
Selection, phenotypical 58Ч60
Selection, random selection on lists 351
Selection, rank-based 164Ч166
Selection, single-step 7
Selection, surviving the GA selection filter 203Ч204
Self Ч Reproduction special issue of Artificial Life journal 469
Selfish Gene, The 52
Sexual reproduction as GA principle 80Ч81
Sierpinski triangle 475
Silent mutations 1 17
Similarity, calculating between generated sets and reference structure 477Ч478
Similarity, Hamming distance for measuring 8
Similarity, measure for strings 14Ч16
Simple Genetic Algorithm, The 208
Simulated diploidy 99Ч101
Simulated diploidy, all possible allele combinations 100
Simulated diploidy, dominance and recessivity 99Ч100
Simulated diploidy, dominance relation 100
Simulated diploidy, extraction of dominant alleles 100Ч101
Simulated diploidy, table of dominant alleles 101
Simulated mimesis of butterflies see butterfly mimesis simulation
Simulating Society 469
Sinc function 267
Single-step selection 7
Sipper, Moshe 469
Smith, John Maynard 52
States of FSA, adding 306Ч307 308
States of FSA, changing the initial state 304Ч305
States of FSA, deleting 307Ч309
States of FSA, final states 300
States of FSA, initial state 299Ч300
States of FSA, modifying the set of states 305Ч309
States of FSA, signals and 299
States, adaptations as 4Ч5
Steady state GA 169
Stochastic L-systems 474
String evolution example, analyzing the first string evolution 24
String evolution example, as "key experiment" 5Ч6
String evolution example, best individual 20
String evolution example, coarse adjustment vs. fine tuning 24
| String evolution example, comparison of three-string evolutions 32Ч33
String evolution example, conservative parameter settings 25
String evolution example, decreased adaptation rate in final phase 24
String evolution example, evolution loop 21
String evolution example, Evolvica implementation 12Ч21
String evolution example, experiments 21Ч33
String evolution example, Hamming distances 8 9 10 20 25 27 28 32
String evolution example, illustrations of string evolution 22Ч23 28Ч30 32
String evolution example, increased mutation radius, effects of 25Ч28
String evolution example, increased mutation, effects of 28Ч32
String evolution example, initial population 16
String evolution example, mutation function 10Ч12
String evolution example, mutation radius 11Ч12 17 18Ч19
String evolution example, mutation rate 11 16Ч17 18Ч19
String evolution example, next generation 19Ч20 21
String evolution example, number decoding for strings 13Ч14
String evolution example, number encoding for strings 8Ч9 13
String evolution example, objective string 6
String evolution example, random string generation 16
String evolution example, selection and mutation algorithm 9Ч12
String evolution example, selection and mutation scheme 7Ч9
String evolution example, similarity measure for strings 14Ч16
String evolution example, single-step selection 7
String evolution example, starting a string evolution 22
String evolution example, string alphabet 6
String evolution example, task description 6
String evolution example, variation vector 16Ч17
String evolution illustrations, mutation radius: 2, mutation rate: 0.1 22Ч23 25 32
String evolution illustrations, mutation radius: 2, mutation rate: 0.2 28Ч31 32
String evolution illustrations, mutation radius: 4, mutation rate: 0.1 26Ч27 32
Structure space, genotypical 58Ч60
Structures as adaptive system components 60 61
Subtree mutation operator (GP) 431
Successor in L-systems 451
Superindividuals, problem of 164Ч165
Survival, adaptation as crucial factor for 5
Survival, GP evolution of balanced mobiles 390
Survival, implicit goals defined by 42 45
Survival, random 162
Survival, schema theorem for GA and 202Ч206
Symbolic expressions see genetic programming on symbolic expressions
Szilard, A. L. 458
Table L-systems 474
Term structures see genetic programming on symbolic expressions
Terms as tree structures 347Ч348
Theory of Evolution, The 52
TIERRA system 294
Time requirements for single-step selection 7
Todd, Stephen 53
Toth, Z. 487
Tournament selection (FSA) 323Ч325
Transactions on Evolutionary Computation 76
Transcription phase of cellular automata 446
Transitions in FSA 299
Transitions in FSA, adding 309Ч311
Transitions in FSA, changing input symbol for 313Ч314 315
Transitions in FSA, changing output symbol for 314Ч316
Transitions in FSA, changing source for 316Ч318
Transitions in FSA, changing target for 318Ч320
Transitions in FSA, deleting 311Ч313
Transitions in FSA, modifying the set of transitions 309Ч313
Transitions in FSA, modifying transitions 313Ч320
Transitions in FSA, network for added states 306Ч307
Translation phase of cellular automata 446
Tree structures, L-systems 466 467 468
Tree structures, symbolic expressions as 365Ч366
Tree structures, terms as 347Ч348
Tree-structured chromosomes 346Ч347
Triple sine (ES multimodal test function) 266Ч268
Triplet encoding of amino acids 88Ч90
Turing machine universal constructor 443
Turn turtle move operation 462
Turtle 462 463
Turtle drawing tool example (see also Lindenmayer systems)
Turtle drawing tool example,  function 459 464 function 459 464
Turtle drawing tool example, changing turtle orientation 460 462
Turtle drawing tool example, changing turtle position 459Ч460
Turtle drawing tool example, drawing graphical elements 462
Turtle drawing tool example, energetic turtle 519
Turtle drawing tool example, environmentally sensitive turtle interpretation 519
Turtle drawing tool example, Hilbert curve demo 463Ч465
Turtle drawing tool example, macro turtle commands 490Ч491
Turtle drawing tool example, modeling of branching structures 465Ч467 468
Turtle drawing tool example, position and orientation of a turtle 458 459
Turtle drawing tool example, turtle interpretation 458 519
Turtle drawing tool example, variation of attributes 462Ч465
Turtle, attributes 462Ч465
Turtle, interpretation 458 519
Typed GP 366Ч367
Uncertain Programming 208
Uniformly distributed random variables 227
Universal constructors 443
Unweaving the Rainbow 52
Update rules for cellular automata 441Ч442
V nyi, R. 487 nyi, R. 487
Variability 57Ч58
Variation vector 16Ч17
Variation, as adaptation step 63
Variation, genotypical 58Ч60
Variation, in evolutionary parameter optimization scheme 73
Videos on genetic programming 396
Visual Models of Plants Interacting with Their Environment 525
Vit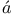 nyi, P. 486 nyi, P. 486
Vogel, Sebastian 3
von Neumann neighborhood for cellular automata 441
von Neumann, John 80 443
Vose, Michael 208
Walsh, Michael 288 298 344
Wellin, Paul 469
Wildwood: The Evolution of L-System Plants for Virtual Environments 526
Wilson, S. 292
Wirth, Niklaus 396
Wolfram, Stephen 440 467
Word sequence for D0L-systems 450
Yaw turtle move operations 461
Young, David 3 52
|
|
 |
| –еклама |
 |
|
|
 |
|
ќ проекте
|
|
ќ проекте



 reduction rules
reduction rules 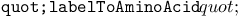 rules
rules 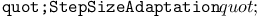 rule
rule 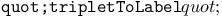 rules
rules  function
function  function
function  function
function  function
function  function
function  nyi, R.
nyi, R.