|
|
 |
| јвторизаци€ |
|
|
 |
| ѕоиск по указател€м |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Jacob C. Ч Illustrating Evolutionary Computation with Mathematica |
|
|
 |
| ѕредметный указатель |
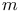 -ploid GA chromosomes see polyploid GA chromosomes -ploid GA chromosomes see polyploid GA chromosomes
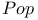 commands for Lindenmayer systems 466 commands for Lindenmayer systems 466
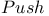 commands for Lindenmayer systems 465Ч466 commands for Lindenmayer systems 465Ч466
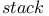 commands for Lindenmayer systems 465Ч466 commands for Lindenmayer systems 465Ч466
 system 469 system 469
 function 307 308 function 307 308
 function 310Ч311 314 function 310Ч311 314
 command ( command ( ) 401 402 404 ) 401 402 404
 command ( command ( ) 402Ч403 404 ) 402Ч403 404
 example 399Ч427 example 399Ч427
 example, analysis of best programs 412Ч417 example, analysis of best programs 412Ч417
 example, ant actions and interpretation of control programs 401 404 406 example, ant actions and interpretation of control programs 401 404 406
 example, avoiding walls 412 413 416 example, avoiding walls 412 413 416
 example, Bernoulli experiment 423Ч427 example, Bernoulli experiment 423Ч427
 example, command sequences 404 example, command sequences 404
 example, comparison of best-evolved programs 422 423 example, comparison of best-evolved programs 422 423
 example, comparison of evolution runs 423Ч427 example, comparison of evolution runs 423Ч427
 example, difficulty of task 401 example, difficulty of task 401
 example, discovering walk-throughs 409 413 415Ч416 example, discovering walk-throughs 409 413 415Ч416
 example, evaluation 405Ч406 example, evaluation 405Ч406
 example, evolution of best individuals 409 417 example, evolution of best individuals 409 417
 example, evolution of program depths 419Ч420 example, evolution of program depths 419Ч420
 example, evolution of the gene pool 418 example, evolution of the gene pool 418
 example, fitness evolution and genome complexity 418Ч420 example, fitness evolution and genome complexity 418Ч420
 example, following food paths 409 412 414 416 422 423 example, following food paths 409 412 414 416 422 423
 example, generating program genomes 404 405 406 407 example, generating program genomes 404 405 406 407
 example, genetic operators 407Ч408 420 422 425 example, genetic operators 407Ч408 420 422 425
 example, influence of operator adaptation 420Ч422 example, influence of operator adaptation 420Ч422
 example, intron regions 402Ч403 example, intron regions 402Ч403
 example, John Muir Trail for 400 example, John Muir Trail for 400
 example, multioperator (MO) GP evolution 425Ч426 example, multioperator (MO) GP evolution 425Ч426
 example, number of ant programs needed 425 example, number of ant programs needed 425
 example, number of evolution runs needed 423Ч424 example, number of evolution runs needed 423Ч424
 example, number of individuals needed 423 424Ч425 example, number of individuals needed 423 424Ч425
 example, perfect foraging 415 416Ч417 example, perfect foraging 415 416Ч417
 example, problem description 401 example, problem description 401
 example, program complexity 419 example, program complexity 419
 example, recombination importance 420 example, recombination importance 420
 example, reduction rules 404 406 example, reduction rules 404 406
 example, repertoire of ant actions 401 example, repertoire of ant actions 401
 example, reproduction-crossover (RC) evolution 425Ч426 example, reproduction-crossover (RC) evolution 425Ч426
 example, response to environmental signals 402 example, response to environmental signals 402
 example, selection templates 407Ч408 example, selection templates 407Ч408
 example, sensors 401 402 412 example, sensors 401 402 412
 example, specialization vs. generalization 422 example, specialization vs. generalization 422
 example, symbolic expression advantages 408 example, symbolic expression advantages 408
 example, time limit 402 example, time limit 402
 example, turning off from walls 412Ч413 415 example, turning off from walls 412Ч413 415
 example, two-dimensional environment for 400 401 example, two-dimensional environment for 400 401
 example, walking along walls 414 416 example, walking along walls 414 416
 package 300 package 300
 function 328Ч329 function 328Ч329
 turtle move operation 460 turtle move operation 460
 function 383Ч384 function 383Ч384
 function 313Ч314 315 function 313Ч314 315
 function 315Ч316 function 315Ч316
 function 317Ч318 function 317Ч318
 function 319 320 function 319 320
 function 227 function 227
 function 95 function 95
 function 120 147 function 120 147
 function, for binary multirecombination 132 function, for binary multirecombination 132
 function, for haploid GA chromosome recombination 136Ч137 function, for haploid GA chromosome recombination 136Ч137
 function for diploid GA chromosomes 97Ч98 104 function for diploid GA chromosomes 97Ч98 104
 function for haploid GA chromosome recombination 136 function for haploid GA chromosome recombination 136
 function for mutation on haploid chromosomes 107 function for mutation on haploid chromosomes 107
 function for polyploid GA chromosomes 98Ч99 function for polyploid GA chromosomes 98Ч99
 function for RNA chromosomes 91 93 function for RNA chromosomes 91 93
 function for binary GA chromosomes 86 87 function for binary GA chromosomes 86 87
 function for real-number GA chromosomes 95 function for real-number GA chromosomes 95
 turtle attribute 463 turtle attribute 463
 turtle attribute 462 turtle attribute 462
 function 199Ч202 function 199Ч202
 function 86 function 86
 function 98 function 98
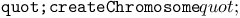 function, for diploid GA chromosomes 97Ч98 function, for diploid GA chromosomes 97Ч98
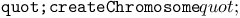 function, for ES chromosomes 214 215 223Ч224 227Ч228 229Ч230 264 272 function, for ES chromosomes 214 215 223Ч224 227Ч228 229Ч230 264 272
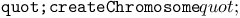 function, for GA chromosomes 84 85 87 function, for GA chromosomes 84 85 87
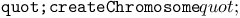 function, for polyploid GA chromosomes 98Ч99 function, for polyploid GA chromosomes 98Ч99
 function, for RNA chromosomes with real-value alleles 94 function, for RNA chromosomes with real-value alleles 94
 function, in GA evolution 172Ч173 function, in GA evolution 172Ч173
 function 386 function 386
 function 16 function 16
 function 127 function 127
 function 13Ч14 function 13Ч14
 function 176 function 176
 function 309 function 309
 function 312Ч313 314 function 312Ч313 314
 function 154 function 154
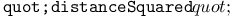 function, string evolution example 14Ч16 function, string evolution example 14Ч16
 function 102Ч103 104Ч105 function 102Ч103 104Ч105
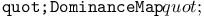 function 100Ч101 102 function 100Ч101 102
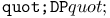 turtle drawing operation 462 turtle drawing operation 462
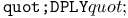 turtle drawing operation 462 turtle drawing operation 462
 function 156Ч157 function 156Ч157
 function 13 function 13
 function 329Ч331 function 329Ч331
 function, comma EP-strategy 331 function, comma EP-strategy 331
 function, comma Ч ES strategy 274 function, comma Ч ES strategy 274
 function, comma Ч GP strategy 374 376 function, comma Ч GP strategy 374 376
 function, decoding and evaluating genotypes 174Ч176 function, decoding and evaluating genotypes 174Ч176
 function, EP evolution experiment 326Ч328 331Ч332 function, EP evolution experiment 326Ч328 331Ч332
 function, ES evolution control function 261Ч263 function, ES evolution control function 261Ч263
 function, ES evolution experiment 264Ч265 269 function, ES evolution experiment 264Ч265 269
 function, GP evolution experiment 374Ч377 function, GP evolution experiment 374Ч377
 function, GP evolution of balanced mobiles 389Ч390 function, GP evolution of balanced mobiles 389Ч390
 function, plus Ч EP strategy 331 function, plus Ч EP strategy 331
 function, plus Ч ES strategy 277 function, plus Ч ES strategy 277
 function, plus Ч GP strategy 374 376 function, plus Ч GP strategy 374 376
 function, starting an ES meta-evolution experiment 271 function, starting an ES meta-evolution experiment 271
 function, starting GP evolution experiments 374 function, starting GP evolution experiments 374
 function, under variable environmental conditions 185Ч190 191Ч192 function, under variable environmental conditions 185Ч190 191Ч192
 function 21 function 21
 function 368Ч370 function 368Ч370
 function, GP term recombination 363 function, GP term recombination 363
 function, recombination at root position 361Ч362 function, recombination at root position 361Ч362
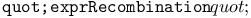 function, recombination of two terms 361 362 function, recombination of two terms 361 362
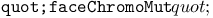 function 120 function 120
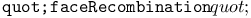 function 147 function 147
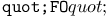 turtle move operation 459 491 turtle move operation 459 491
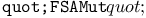 function, function,  function 307 308 function 307 308
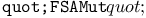 function, function,  function 310Ч311 314 function 310Ч311 314
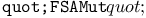 function, function, 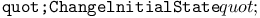 function 304Ч305 function 304Ч305
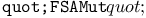 function, function, 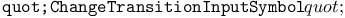 function 313Ч314 315 function 313Ч314 315
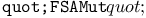 function, function, 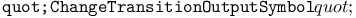 function 315Ч316 function 315Ч316
 function, function,  function 317Ч318 function 317Ч318
 function, function,  function 319 320 function 319 320
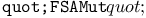 function, function,  function 309 function 309
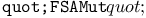 function, function, 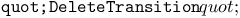 function 312Ч313 314 function 312Ч313 314
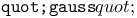 function, function, 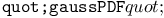 function 219Ч220 function 219Ч220
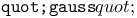 function, for step size adaptation 229 230 function, for step size adaptation 229 230
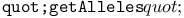 function 116 function 116
 turtle drawing operation 462 turtle drawing operation 462
 commands ( commands ( ), command stack 403 ), command stack 403
 commands ( commands ( ), implicit loop 403 ), implicit loop 403
 commands ( commands ( ), overview 402 ), overview 402
 commands ( commands ( ), selection templates 407Ч408 ), selection templates 407Ч408
 commands ( commands ( ), symbolic expression advantages for 408 ), symbolic expression advantages for 408
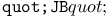 turtle move operation 460 turtle move operation 460
 turtle move operation 460 turtle move operation 460
 turtle move operation 460 turtle move operation 460
 turtle move operation 460 turtle move operation 460
 rules 89 90 rules 89 90
 expressions 474 expressions 474
 expressions 474 expressions 474
 command 300 command 300
 function 320 function 320
 function 321Ч322 function 321Ч322
 function 321 function 321
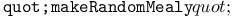 function 320Ч321 function 320Ч321
|  function 388Ч389 function 388Ч389
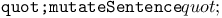 function 16Ч19 function 16Ч19
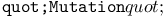 function (see also function (see also 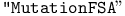 function) function)
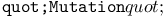 function, for binary haploid GA chromosomes 108 function, for binary haploid GA chromosomes 108
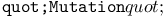 function, for ES chromosomes (without step size adaptation) 223 function, for ES chromosomes (without step size adaptation) 223
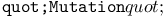 function, for ES chromosomes with step size adaptation 226 function, for ES chromosomes with step size adaptation 226
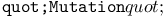 function, for GP chromosomes 370Ч371 function, for GP chromosomes 370Ч371
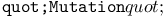 function, for polyploid GA chromosomes 109Ч110 function, for polyploid GA chromosomes 109Ч110
 function 304Ч305 (see also function 304Ч305 (see also 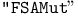 function) function)
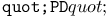 turtle move operation 461 turtle move operation 461
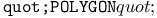 turtle drawing operation 462 turtle drawing operation 462
 function 328Ч330 function 328Ч330
 turtle move operation 461 turtle move operation 461
 function, generating GP terms with variable numbers of arguments 355 356 function, generating GP terms with variable numbers of arguments 355 356
 function, GP term mutation 368Ч369 function, GP term mutation 368Ч369
 function, term composition on a set of functions and terminals 350Ч351 function, term composition on a set of functions and terminals 350Ч351
 function 147 function 147
 function 236Ч237 function 236Ч237
 function 351 function 351
 function, for global recombination 240Ч241 function, for global recombination 240Ч241
 function, for local recombination 236Ч237 function, for local recombination 236Ч237
 dominance relation 101 dominance relation 101
 function, ES recombination overview 243Ч245 function, ES recombination overview 243Ч245
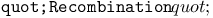 function, for global multi Ч ES recombination 250 function, for global multi Ч ES recombination 250
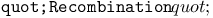 function, for haploid GA chromosomes 135Ч136 function, for haploid GA chromosomes 135Ч136
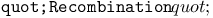 function, for local binary ES recombination 246Ч247 function, for local binary ES recombination 246Ч247
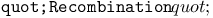 function, for local multi Ч ES recombination 248Ч249 function, for local multi Ч ES recombination 248Ч249
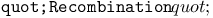 function, for multiple global and local ES recombination 251 function, for multiple global and local ES recombination 251
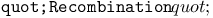 function, for triploid GA chromosomes 142 function, for triploid GA chromosomes 142
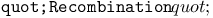 function, GA recombination overview 134Ч135 function, GA recombination overview 134Ч135
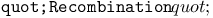 function, GP recombination overview 364Ч365 function, GP recombination overview 364Ч365
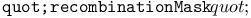 function 129 function 129
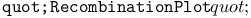 function 242Ч243 function 242Ч243
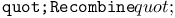 function, for binary componentwise GA recombination 124Ч126 function, for binary componentwise GA recombination 124Ч126
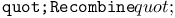 function, for binary multi Ч GA recombination 132 function, for binary multi Ч GA recombination 132
 function, for crossover of nonhomologous GA chromosomes 159 function, for crossover of nonhomologous GA chromosomes 159
 function, for discrete componentwise multi Ч GA recombination 126 function, for discrete componentwise multi Ч GA recombination 126
 function, for ES multirecombination on parameter lists 233Ч234 function, for ES multirecombination on parameter lists 233Ч234
 function, for four-range multi-GA recombination 132Ч133 function, for four-range multi-GA recombination 132Ч133
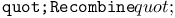 function, for GA four-point crossover 128 function, for GA four-point crossover 128
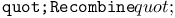 function, for GA one-point crossover 127 function, for GA one-point crossover 127
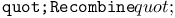 function, for global ES recombination 241 function, for global ES recombination 241
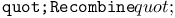 function, for masked GA multi-point crossover 130Ч131 function, for masked GA multi-point crossover 130Ч131
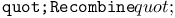 function, for masked GA one-point crossover 130 function, for masked GA one-point crossover 130
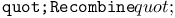 function, for masked GA recombination by example 129 function, for masked GA recombination by example 129
 function 92 93 function 92 93
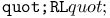 turtle move operation 461 turtle move operation 461
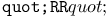 turtle move operation 461 turtle move operation 461
 function 452Ч453 function 452Ч453
 function 194Ч196 function 194Ч196
 function 264 function 264
 function 374 375Ч376 function 374 375Ч376
 command 300 command 300
 function, for mutated ES chromosomes 224 function, for mutated ES chromosomes 224
 function, for mutated ES chromosomes with adapted step sizes 228 230 function, for mutated ES chromosomes with adapted step sizes 228 230
 function, overview 214Ч215 function, overview 214Ч215
 function, for unbalanced mobiles 281Ч282 function, for unbalanced mobiles 281Ч282
 function, overview 378Ч379 function, overview 378Ч379
 function, visualizing mobiles 379Ч380 function, visualizing mobiles 379Ч380
 function, for unbalanced mobiles 281Ч282 function, for unbalanced mobiles 281Ч282
 function, overview 378Ч379 function, overview 378Ч379
 function, visualizing mobiles 380Ч381 function, visualizing mobiles 380Ч381
 function 387 function 387
 function 387 function 387
 function 249 function 249
 function 141Ч142 function 141Ч142
 rule 226 227 rule 226 227
 command ( command ( ) 402Ч403 404 ) 402Ч403 404
 function 182 function 182
 function 265 269 function 265 269
 turtle attribute 463 turtle attribute 463
 function 353Ч354 355 363 function 353Ч354 355 363
 turtle attribute 463 turtle attribute 463
 turtle attribute 463 turtle attribute 463
 turtle move operation 462 turtle move operation 462
 rules 89 rules 89
 turtle attribute 463 turtle attribute 463
 command ( command ( ) 401 402 404 ) 401 402 404
 command ( command ( ) 401 402 404 ) 401 402 404
 function 181 function 181
 turtle move operation 461 turtle move operation 461
 turtle move operation 461 491 turtle move operation 461 491
Accommodation phase of FSA 302
Adami, Christoph 469
Adaptation in Natural and Artificial Systems 79 208
Adaptations, adaptation steps 62Ч63
Adaptations, as crucial factor for survival 5
Adaptations, as processes 5
Adaptations, as states 4Ч5
Adaptations, components of adaptive systems 61
Adaptations, defined 79
Adaptations, ES mutation with step size adaptation 218 225Ч230
Adaptations, gene pool diversity and 190
Adaptations, mimesis 33Ч34
Adaptations, of individuals vs. populations 4Ч5
Adaptations, operator weight adaptation in GP 428Ч430
Adaptations, progressive change of structures 60
Adaptive systems, feedback components 60
Adaptive systems, main components 61
Adaptive systems, neural networks as 61
Adding, FSA states 306Ч307 308
Adding, FSA transitions 309Ч311
Advances in Genetic Programming 397
AI see artificial intelligence (AI)
AL see artificial life (AL)
Algorithmic Beauty of Plants, The 524
Algorithms (see also evolutionary algorithms for optimization; genetic algorithms)
Algorithms, adaptation algorithm 61
Algorithms, evolutionary algorithms 344 399
Algorithms, evolutionary parameter optimization scheme 71Ч75
Algorithms, for GA evolution 171Ч173
Algorithms, for GA with Evolvica 169Ч173
Algorithms, for selection and mutation of strings 9Ч12
Algorithms, general evolutionary algorithm scheme 63Ч75
Algorithms, induction of programs by evolutionary algorithms 285
Algorithms, of evolution as reproductive plan 64 65
Alleles 85Ч86
Alleles, alphabet setting in Evolvica 85
Alleles, binary GA chromosome with 15
Alleles, dominance and recessivity 99Ч101
Alleles, extraction from RNA chromosomes 116
Alleles, extraction of dominant alleles 100Ч105
Alleles, GA chromosome interpretation 101Ч105
Alleles, point mutation and diversity of 105
Alleles, point mutation of homologous alleles 110
Alleles, possible combinations for diploid chromosomes 100
Alleles, real-value, chromosomes with 93Ч95
American Standard Code for Information Interchange (ASCII) 8
Anabaena catenula growth simulation 451Ч453
Angeline, P. J. 435
Architecture-altering operations (GP) 432Ч434
Architecture-altering operations (GP), automatically defined functions 432Ч433
Architecture-altering operations (GP), automatically defined iterations and loops 434
ArtFlower garden 490Ч493 514Ч518
ArtFlower garden, comparing growth dynamics 517
ArtFlower garden, evolutionТs creativity 514Ч516
ArtFlower garden, genetic operators 497
ArtFlower garden, growth and recursive branching 492 493 494 495
ArtFlower garden, L-system breeding 514
ArtFlower garden, L-system evaluation 514
ArtFlower garden, L-systems of flowering plant 490 491
ArtFlower garden, Lychnis coronaria 490
ArtFlower garden, macro turtle commands 490Ч491
ArtFlower garden, structure formation and growth 492
ArtFlower garden, structure formation dynamics 514
Artificial intelligence (AI) (see also evolutionary programming)
Artificial intelligence (AI), bottom-up approach 297 549
Artificial intelligence (AI), EP approach 297Ч298
Artificial intelligence (AI), evolution and 3
Artificial intelligence (AI), heuristic programming 297
|
|
 |
| –еклама |
 |
|
|
 |
|
ќ проекте
|
|
ќ проекте



 -ploid GA chromosomes
-ploid GA chromosomes  commands for Lindenmayer systems
commands for Lindenmayer systems  commands for Lindenmayer systems
commands for Lindenmayer systems  commands for Lindenmayer systems
commands for Lindenmayer systems  system
system  function
function  function
function  command (
command ( )
)  command (
command ( package
package  function
function  turtle move operation
turtle move operation  function
function  function
function  function
function  function
function  function
function  function
function  function
function  function
function  function, for binary multirecombination
function, for binary multirecombination  function for diploid GA chromosomes
function for diploid GA chromosomes  function for binary GA chromosomes
function for binary GA chromosomes  turtle attribute
turtle attribute  turtle attribute
turtle attribute  function
function  function
function  function
function  function, for diploid GA chromosomes
function, for diploid GA chromosomes  function
function  function
function  function
function 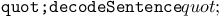 function
function  function
function  function
function 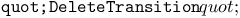 function
function  function
function 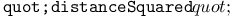 function, string evolution example
function, string evolution example  function
function 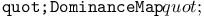 function
function 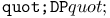 turtle drawing operation
turtle drawing operation 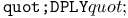 turtle drawing operation
turtle drawing operation  function
function  function
function  function
function  function, comma EP-strategy
function, comma EP-strategy  function
function  function
function  function, GP term recombination
function, GP term recombination  function
function  function
function  turtle move operation
turtle move operation 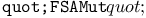 function,
function, 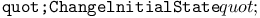 function
function 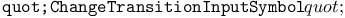 function
function 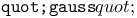 function,
function, 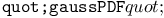 function
function 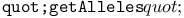 function
function  turtle drawing operation
turtle drawing operation  commands (
commands ( turtle move operation
turtle move operation  turtle move operation
turtle move operation  turtle move operation
turtle move operation  turtle move operation
turtle move operation  rules
rules  expressions
expressions  expressions
expressions  command
command  function
function  function
function  function
function  function
function 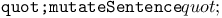 function
function 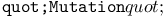 function
function 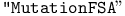 function)
function) function
function 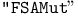 function)
function) turtle move operation
turtle move operation  turtle drawing operation
turtle drawing operation  function
function  turtle move operation
turtle move operation  function, generating GP terms with variable numbers of arguments
function, generating GP terms with variable numbers of arguments 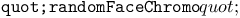 function
function  function
function 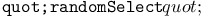 function
function 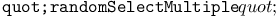 function, for global recombination
function, for global recombination 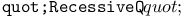 dominance relation
dominance relation  function, ES recombination overview
function, ES recombination overview  function
function  function
function  function, for binary componentwise GA recombination
function, for binary componentwise GA recombination  function
function  turtle move operation
turtle move operation  turtle move operation
turtle move operation  function
function  function
function  function
function  function
function  command
command  function, for mutated ES chromosomes
function, for mutated ES chromosomes  function, for unbalanced mobiles
function, for unbalanced mobiles  function, for unbalanced mobiles
function, for unbalanced mobiles  function
function  function
function  function
function  function
function  rule
rule  command (
command ( function
function 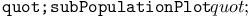 function
function  turtle attribute
turtle attribute 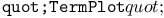 function
function  turtle attribute
turtle attribute  turtle attribute
turtle attribute 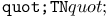 turtle move operation
turtle move operation 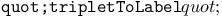 rules
rules  turtle attribute
turtle attribute  command (
command ( )
)  command (
command ( function
function  turtle move operation
turtle move operation  turtle move operation
turtle move operation