|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Frenkel D., Smit B. — Understanding Molecular Simulation: from algorithms to applications |
|
|
 |
| Предметный указатель |
Harmonic oscillator, trajectories 156 158
Harris, J. 271 272 331
Harvey, A.H. 223
Hasslacher, B. 476
Hautman,J. 317
Haymet, A.D.J. 236
Hcp, free energy 261
heat capacity 58 85
Heermann, D.W. 6 398
Hegger,R. 283
Heinbruch,U. 178
Hellekalek, P. 588
Helmholtz free energy 116
Helmholtz free energy, definition 12
Helmholtz free energy, elastic constants 520
Helmholtz free energy, excess 116
Helmholtz free energy, ideal gas 116
Henderson, D. 236 243 318
Hendriks, E.M. 222
Henkelman, G. 463
Henry coefficient 280
Hermann, D.W. 125 217
Heyes,D.M. 316 317
Hilbers, PA.J. 362 403
Histogram reweighting technique 395
Histogram reweighting technique, example 394
Hiwatari, Y. 167
Hockney, R.W. 6 75 292 310 311 314 550
Hoefsloot, H.C.J. 471 474
Holm,C 311 312 314
Holmes, S. 55
Holt,A.C. 522 523
Hoogerbrugge, P.J. 466 469
Hoover, W.G. 6 63 147 152 156 159 235 236 242 243 256 261 510 516 522 523
Hoye,J.S. 222
Huse,D.A. 261 263
Hut, P. 306
Hydrodynamics, example 474
Hyper-parallel tempering 395
Hyperdynamics 464
Ideal chain 276 337 366
Ideal chain, chemical potential 368
Iedema,PD. 474
Ilario,G. 585
Impey,R.W 523
Importance-sampling scheme 24
Initialization, algorithm 66
Interfacial tension 472
Internal potential energy 276
Interstitial, concentration 263
Ion, example 460
Irving, J.H. 472
Ising model, finite-size effects 219
Ising, exercise 137
Isobaric-isothermal ensemble, case study 122 123
Isobaric-isothermal ensemble, Monte Carlo technique 115
Isobaric-isothermal ensemble, schematic sketch 117
Isotension-isothermal ensemble, Monte Carlo technique 125
Jackson, G. 136 209
Jacobian 490
Jacobian, elastic constant 521
Jacobson, J.D. 6 235 237
Jacucci,G. 529
Jarzynski, C. 196
Jarzynski’s identity 196
Joannopoulos, J.D. 501
Johnson, J.K. 52 53 54 55 57 123 133 145 146 213 231
Johnson, K.W. 236 243
Jonsson, H. 462 463
Jorgensen, W.L. 372 373
June,R.L. 135
Kalikmanov, V.I. 222
Kalos,M.H. 6 30
Kantor,Y. 523
Kapral,R. 466 478
Karaborni, S. 362 368 372 373 374
Karasawa, N. 306 315
Karavias, F. 135
Kennedy, A. 398
Kerkhof, P.J.A.M. 582
Kirkpatrick, S. 389
Kirkwood g factor 302
Kirkwood — Buff 472
Kirkwood,J.G. 170 472
Kirkwood’s coupling parameter method 170
Klein, M.L. 106 157 159 317 423 424 427 503 523 535 536 537 538 543 544 584 585
Kleinman, L. 501
Koelman, J.M.V.A. 466 469
Kofke, D.A. 168 202 225 229 231 233 234 236 237 238 239 241
Kolafa,J.A. 304 389
Koonin, S.E. 6
Koper,G.J.M. 117
Koutras,N.K. 223
Kramer, H.A. 445
Kranendonk, W.G.T. 43 171 227
Kremer, K. 280 281 283 331
Krishna, R. 351 352 362 370 383
Kron,A.K. 4
Kumar, S.K. 270 350 374
Ladd, A.J.C. 98 167 171 236 243 244 245 246 258 260 261 262 466 477 522
Lagrangian 481
Lagrangian strain tensor 519
Lagrangian, example 485
Laird, B.B. 236
Landau, D.P 6 167
Laso,M. 271 331 372
Lattice Boltzmann method 476
Lattice gas cellular automata 476
Lattice-coupling-expansion method 246
Lattice-switch Monte Carlo 262
Law of rectilinear diameters 217
Leap Frog algorithm 75
Lebowitz, J.L. 84 111 178
Lee,H. 311 313 314
Lee,M.A. 306 310 315
Leighton, R.B. 481
Lekkerkerker, H.N.W. 171 234
Lennard — Jones chains, case study 340 382
Lennard — Jones chains, equation of state 340 342
Lennard — Jones chains, recoil growth 382
Lennard — Jones, algorithm force calculation 68
Lennard — Jones, Andersen thermostat 142
Lennard — Jones, case study 51 54 56 98 100 101 122 123 133 142 153 175 181 211
Lennard — Jones, chemical potential 133 175 177 181 182
Lennard — Jones, diffusion 100—102
Lennard — Jones, energies 99
Lennard — Jones, equation of state 51 53 55 57 122 123 133 146
Lennard — Jones, example 394
Lennard — Jones, exercise 60 105
Lennard — Jones, finite-size effects 220
Lennard — Jones, force 69
Lennard — Jones, Gibbs ensemble 214
Lennard — Jones, mean-squared displacement 102 147 155
Lennard — Jones, molecular dynamics 98 100
Lennard — Jones, Nose — Hoover thermostat 153
Lennard — Jones, phase diagram 38 214
Lennard — Jones, phase equilibria 123
Lennard — Jones, radial distribution function 101
Lennard — Jones, statistical error, calculation of 100
Lennard — Jones, truncated and shifted potential 98
Lennard — Jones, truncation of the potential, effect of 38
Lennard — Jones, vapor-liquid coexistence 124
Lennard — Jones, velocity autocorrelation function 102
Lennard — Jones, velocity distribution 145 154
LeSar, R.A. 171 243 244 245 246
Levesque,D. 222 292
Levy,R.M. 316
Limcharoen, P. 134
Linear response theory 509
Linear response theory, dissipation 513
| Linear response theory, dynamic 511
Linear response theory, static 509
Linked lists 550
Linked lists, algorithm 551—553
Liouville formulation, multiple time step 424
Liouville formulation, Nose — Hoover algorithm 536
Liouville operator 78
Liouville theorem, non-Hamiltonian system 496
Liu, Yi 156 495 497 507
Lofti, A. 213
Long-range interactions 36
Long-range interactions, example 314
Lowe, C.P. 312 467 477
Lowen, H. 422
Loyens, L.D.J.C. 361
Lustig,R. 176
Luty, B.A. 315
Lutz,C. 551
Lyapunov instability 81
Lyubartsev, A.P 389
Macedonia, M.D. 345
MacGowan, D. 135
Machlup,S. 84
Mackie,A.D. 473 474
Madden, PA. 171 422 423
Madden, T.J. 474 475
Maddox,M.W. 135
Madura, J.D. 372 373
Maesen, T.L.M. 368
Maginn,E.J. 281 282 345
Malevanets, A. 466 478
MandeLM.J. 35
Manousiouthakis, VI. 42
MarchLM. 427 428
Marinari, E. 389
Markov chain 29
Marsh, C.A. 469 473
Martin del Rio, E. 220 223
Martin, M.G. 352 353 362 374 383
Martsinovski, A.A. 389
Martyna, G.J. 77 106 156 157 159 160 398 409 424 427 428 495 497 503 507 535 536 537 538 543 544 584 585
Massobrio, C. 176 178 226
Masters, A.J. 469
Matrix, antisymmetric 490
Matrix, Jacobian 490
Matrix, symplectic 491
Matrix, transposed 491
Matsuda,H. 236 243
Mau,S.-C. 261 263
Mavrantzas, V.G. 353 358 360
Maxwell — Boltzmann distribution 66
McCabe,C. 374
McCormick, A V. 135
McDonald, I.R. 3 4 84 90 111 116 119 178 195 509 582
McNamara, G.R 476 477
Mean-squared displacement, algorithm 91 95
Mehlig, B. 398
Meijer, E.J. 171 234 243 244 245 246
Meirovitch, H. 287 375
Meiss,J.D. 494
Meller,J. 84
Mesoscale dynamics 476
Mesoscopic model 465
Methane, adsorption isotherm 135
Methane, example 134
Metropolis scheme, schematic sketch 28
Metropolis, N. 4 24 27 32 174
Meyer, M. 6 63 421
Mezei,M. 128 133 221
Microcanonical ensemble, Monte Carlo technique 114
Microcanonical ensemble, partition function 492
Miiller,M. 270
Miiser,M.H. 443
Milgram, M. 4
Miller, M.A. 55
Miller, W.H. 442
Mills, G. 462
Minimum image convention 39
Mobility 513
Model fluid,  231 231
Model fluid, alkanes 372
Model fluid, block copolymers 474
Model fluid, dipolar hard-sphere fluid 222
Model fluid, hard spheres 237 256 261
Model fluid, ideal chains 345
Model fluid, ions in water 460
Model fluid, Lennard — Jones 51 54 56 98 100 101 122 123 133 142 153 175 181 211 394
Model fluid, Lennard — Jones chains 340
Model fluid, Lennard — Jones dumbbell 427
Model fluid, methane 134
Model fluid, point dipoles 330
Model fluid, polymers 396
Model fluid, restricted primitive model 221
Model fluid, soft spheres 235
Model fluid, Stockmayer fluid 170 222
Model fluid, water 329
Molecular dynamics, algorithm (NVE) 65 66 70
Molecular dynamics, algorithm (NVT) 143 144
Molecular dynamics, boundary conditions 32
Molecular dynamics, case study 98 100 101
Molecular dynamics, exercise 105 161
Molecular dynamics, initialization 40
Molecular dynamics, Lennard — Jones 98 100 101
Molecular dynamics, NPT ensemble 158
Molecular dynamics, NVE ensemble 64
Molecular dynamics, NVT ensemble 140 147 155
Molecular dynamics, potential, truncation of 35
Mon,K.K. 218 219 270
Mondello,M. 374
Monson,P.A. 241
Monte Carlo integration, exercise 59
Monte Carlo technique, algorithm 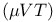 131 132 131 132
Monte Carlo technique, algorithm (fixed center of mass) 251
Monte Carlo technique, algorithm (Gibbs) 209 210 212
Monte Carlo technique, algorithm (NPT) 121 122
Monte Carlo technique, algorithm (NVT) 33
Monte Carlo technique, boundary conditions 32
Monte Carlo technique, canonical ensemble 112
Monte Carlo technique, canonical ensemble: justification of algorithm 114
Monte Carlo technique, case study 51 54 56 122 123 133 211 256
Monte Carlo technique, configurational-bias Monte Carlo (lattice) 334 335
Monte Carlo technique, efficiency 119
Monte Carlo technique, grand-canonical ensemble 126
Monte Carlo technique, initialization 40
Monte Carlo technique, isobaric-isothermal ensemble 115
Monte Carlo technique, isotension-isothermal ensemble 125
Monte Carlo technique, justification 112
Monte Carlo technique, Metropolis scheme 28
Monte Carlo technique, microcanonical ensemble 114
Monte Carlo technique, orientational bias 325
Monte Carlo technique, path ensemble 454
Monte Carlo technique, potential, truncation of 35
Monte Carlo technique, random sampling 24
Monte Carlo, dynamic 31
Monte Carlo, end-bridging 357
Monte Carlo, exercise 59—61 136 137 161
Monte Carlo, rebridging 357
Mooij, G.C.A.M. 234 270 271 282 331 341 372 374 573
Moore, J.D. 374
Morriss,G.P. 6 141
Mountain, R.D. 47 223
Mouritsen, O.G. 6 167
Mousseau, N. 463
Mulder, B.M. 171
Multicanonical method 262
Multiple time step, algorithm 426
Multiple time step, case study 427
Multiple time step, Liouville formulation 424
Mundy,CJ. 495
Myers, A.L. 135
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте