|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Frenkel D., Smit B. — Understanding Molecular Simulation: from algorithms to applications |
|
|
 |
| Предметный указатель |
Abraham, KF. 551
Abramowitz, M. 260
Acceptance rule, biased sampling 323
Acceptance rule, canonical ensemble 29 32 113
Acceptance rule, CBMC fixed endpoints 355
Acceptance rule, configurational-bias Monte Carlo 332 334 339
Acceptance rule, Gibbs ensemble 205
Acceptance rule, Gibbs ensemble technique 372
Acceptance rule, grand-canonical ensemble 130 367
Acceptance rule, isobaric-isothermal ensemble 118
Acceptance rule, Metropolis scheme 29
Acceptance rule, NPT ensemble 118
Acceptance rule, orientational bias 324
Acceptance rule, parallel tempering 390
Acceptance rule, path ensemble 455
Acceptance rule, semigrand ensemble 230
Acceptance-rejection technique 341
Accepting a trial move 30
Ackland, G.J. 261 262 263
Activation-relaxation technique 463
Adams, D.J. 1ll 128 178 257 258
Adiabatic transformation 172
Adolf, D.B. 316 317
Adsorption, example 134
Adsorption, methane in zeolite 135
Agrawal, R. 168 234 236
Ailawadi, N.K. 527 529
Alder, B.J. 4 6 167 235 237 257 263 266 478
Alexander, F.J. 478
Alexandrowicz, Z. 375
Algorithm xvi
Algorithm, cell lists 551—553
Algorithm, cell lists and Verlet lists 554 555
Algorithm, combined lists 554 555
Algorithm, configurational-bias Monte Carlo 344 346 347
Algorithm, configurational-bias Monte Carlo (lattice) 334 335
Algorithm, diffusion 91 95
Algorithm, equations of motion: Andersen thermostat 144
Algorithm, equations of motion: Nose — Hoover thermostat 540—542
Algorithm, equations of motion: Verlet algorithm 70
Algorithm, exchange of particle 132
Algorithm, force, calculation of the 68
Algorithm, Gaussian distribution 579
Algorithm, generate an Einstein crystal 252
Algorithm, generate bond and torsion angle 580
Algorithm, generate bond angle 579
Algorithm, generate bond length with harmonic springs 578
Algorithm, Gibbs ensemble technique 209 210 212
Algorithm, growing a chain on a lattice 335
Algorithm, growing an alkane 344
Algorithm, growing ethane 346
Algorithm, growing propane 346
Algorithm, initialization 66
Algorithm, linked lists 551—553
Algorithm, mean-squared displacement 91 95
Algorithm, Molecular Dynamics: Andersen thermostat 143
Algorithm, Molecular Dynamics: Nose — Hoover thermostat 540—542
Algorithm, Molecular Dynamics: NVE ensemble 65
Algorithm, Monte Carlo technique (NVT) 251
Algorithm, Monte Carlo technique: 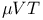 ensemble 131 132 ensemble 131 132
Algorithm, Monte Carlo technique: (fixed center of mass) 251
Algorithm, Monte Carlo technique: NPT ensemble 121 122
Algorithm, Monte Carlo technique: NVT ensemble 33
Algorithm, multiple time step 426
Algorithm, orientational bias 325
Algorithm, particle displacement 33
Algorithm, particle displacement (fixed center of mass) 251
Algorithm, particle exchange (Gibbs) 212
Algorithm, particle insertion method 175
Algorithm, radial distribution function 86
Algorithm, random vector on a unit sphere 578
Algorithm, selection of trial orientations 577
Algorithm, trial position of n-alkane 347
Algorithm, velocity autocorrelation function 91 95
Algorithm, Verlet 82
Algorithm, Verlet lists 547—549
Algorithm, volume change (Gibbs) 210
Algorithm, volume change (NPT) 122
Algorithm, Widom method 175
Alkanes, critical properties 372
Alkanes, example 280 368
Alkanes, generation of trail positions 342
Allen, M.P. 6 30 39 48 49 58 63 85 216 277 397 421 467 510 529 578 584
Amar,J.G. 223
Amon, L.M. 55
Andersen thermostat, algorithm 143 144
Andersen thermostat, case study 142
Andersen thermostat, exercise 161
Andersen thermostat, harmonic oscillator 155
Andersen thermostat, Lennard — Jones 142
Andersen, H.C. 75 125 139 141 142 144 146 147 159 267
Anderson, H.L. 27
Anselme, M.J. 373
Antisymmetric matrix 490
Appel,A.W. 306
Attard, P. 117
Auer,S. 394 396 462
Auerbach, D.J. 551
Azadipour, A.Z. 389
Azhar,F.E. 234
Backx,G. 469 473
Bakker,A.F 472 551
Balian, R. 15
Banaszak, B.J. 374
Barber, M. 316 317
Barkema, G.T. 6 463
Barker, J.A. 4 236 243
Barnes, J. 306
Barrier crossing, case study 440
Bastolla,U. 286
Bates, M.A. 267 523
Batoulis, J. 280 281 283 331
Baus, M. 6
Beckers, J.V.L. 312
Beeman algorithm 76
Bekker,H. 546 550
Bell,A.T. 135 281 282
Bennet — Chandler approach 436
Bennett, C.H. 179 187 189 263 266 432 575
Benzi,R. 476 477
Berendsen, H.J.C. 77 161 162 172 414 427 546 550
Berens,P.H. 75 144
Berg,B.A. 262
Berkowitz, M.L. 311 313 314 317 318
BernaLJ.D. 3 4
Berne, B.J. 77 316 398 409 432 462 584
Biben,T. 361 363
Binder, K. 6 125 167 181 217 218 219 280 331 399 523
Bird,G.A. 477 478
Bladon,P 361
Blander, M. 423
Bolhuis, PG. 202 234 237 239 261 361 363 364 365 450 453 456
Bond formation scheme 405
Bond formation scheme, acceptance rule 405
Bonded potential energy 337
Bonet Avalos, J. 473 474
Boone, T.D. 51 353 358 359 360
boundary conditions 32
Bowles, R.K. 266
Branched alkanes, configurational-bias Monte Carlo 350
Brey,J.J. 439 447 450
Brodka,A. 316 317
Brooks, B.R. 291
Broughton, J.Q. 244
Brown, B.C. 236 243
Brownian dynamics 474
Bruce, A.D. 125 217 261 262 263 395
Bruin, C. 472
Buff,F.P 472
Bunker, A. 394 397
| Caillol,J.-M. 222 292 330
Camp, P.J. 222
Canonical ensemble, , Monte Carlo technique, justification of 114
Canonical ensemble, Monte Carlo technique 112
Canonical transformation, symplectic condition 491
Cape, J.N. 167 236
Car,R. 410 421
Case study xvi
Case Study, 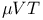 ensemble 133 ensemble 133
Case Study, Andersen thermostat 142
Case Study, barrier crossing 440
Case Study, cell lists 554
Case Study, chemical potential: Lennard — Jones 175 181
Case Study, comparison CPU saving schemes 554
Case Study, configurational-bias Monte Carlo 340 345
Case Study, constraints 427
Case Study, detailed balance 54
Case Study, diffusion 100 101
Case Study, Dissipative particle dynamics 470
Case Study, dynamic properties of the Lennard — Jones fluid 100
Case Study, Einstein crystal 256
Case Study, equation of state: Lennard — Jones 51 122 133
Case Study, equation of state: Lennard — Jones chains 340
Case Study, Gibbs ensemble technique 211
Case Study, hard spheres 256
Case Study, harmonic oscillator 155 157
Case Study, keep old configuration 56
Case Study, Lennard — Jones 51 54 56 98 100 101 122 123 133 142 153 175 181 211
Case Study, Molecular Dynamics 98 100 101
Case Study, Monte Carlo technique 51 54 56 122 123 133 211 256
Case Study, multiple time step 427
Case Study, Nose — Hoover thermostat 153
Case Study, NPT ensemble 122 123
Case Study, NVT ensemble 51
Case Study, overlapping distribution 181
Case Study, parallel tempering 391
Case Study, particle insertion method 175
Case Study, path ensemble 456
Case Study, phase equilibria: Lennard — Jones 123 211
Case Study, rare events 440 456
Case Study, recoil growth 382
Case Study, SHAKE 427
Case Study, solid-liquid phase equilibrium of hard spheres 256
Case Study, static properties of the Lennard — Jones fluid 98
Case Study, trial configurations of ideal chains 345
Case Study, Verlet lists 554
Case Study, Widom method 175
Catlow,C.R.A 134
Cell lists 550
Cell lists, algorithm 551—553
Cell lists, case study 554
Chain molecules, chemical potential 270
Chain molecules, concerted rotation 51
Chain molecules, example 396
Chandler, D. 193 353 403 404 432 443 450 453 456 462 509
Chao,K.C. 128
Chemical potential, acceptance ratio method 189
Chemical potential, case study 175 181
Chemical potential, chain molecules 270
Chemical potential, excess chemical potential 174 211
Chemical potential, finite-size corrections 178
Chemical potential, Gibbs ensemble 211
Chemical potential, ideal gas 129 560
Chemical potential, incremental 270
Chemical potential, Lennard — Jones 175 181
Chemical potential, mixtures 226
Chemical potential, modified Widom method 270
Chemical potential, multiple-histograms 183
Chemical potential, NPT ensemble 177
Chemical potential, NVE ensemble 178
Chemical potential, NVT ensemble 174
Chemical potential, overlapping distribution 179 282
Chemical potential, particle insertion method 173 174
Chemical potential, recursive sampling 283
Chemical potential, Rosenbluth sampling 279
Chemical potential, self-consistent histogram method 184
Chemical potential, tail correction 176
Chemical potential, thermodynamic integration 269
Chemical potential, umbrella sampling 192
Chemical potential, Widom method 173 174
Chen, B. 345 374
Chen,S. 476 477
Chen,Z. 357
Cho,K. 501
Chung, S.T. 178
Ciccotti, G. 3 6 50 51 63 156 176 178 226 253 414 421 427 432 437 440 443 462 495 497 507 510 516
Clarke, J.H.R. 316 317
Clausius — Clapeyron equation 233
Cluster moves, example 403
Coarse-grained model 465
Cochran, H.D. 374
Cohen, L.K. 128
Coker,D.F. 432 462
Colloids 465
Colloids, example 363
Compressibility, phase space 496
Concerted rotation 51 357
Configurational-bias Monte Carlo acceptance rule 332 334 339
Configurational-bias Monte Carlo acceptance rule, algorithm (alkane) 344 347
Configurational-bias Monte Carlo acceptance rule, algorithm (ethane) 346
Configurational-bias Monte Carlo acceptance rule, algorithm (lattice) 334 335
Configurational-bias Monte Carlo acceptance rule, algorithm (propane) 346
Configurational-bias Monte Carlo acceptance rule, branched alkanes 350
Configurational-bias Monte Carlo acceptance rule, case study 340 345
Configurational-bias Monte Carlo acceptance rule, exercise 384 386
Configurational-bias Monte Carlo acceptance rule, explicit-hydrogen model 345
Configurational-bias Monte Carlo acceptance rule, fixed endpoints (continuum) 355
Configurational-bias Monte Carlo acceptance rule, fixed endpoints (lattice) 353
Configurational-bias Monte Carlo acceptance rule, Gibbs ensemble technique 370
Configurational-bias Monte Carlo acceptance rule, justification (lattice) 334
Configurational-bias Monte Carlo acceptance rule, justification (off-lattice) 339
Configurational-bias Monte Carlo acceptance rule, lattice 332
Configurational-bias Monte Carlo acceptance rule, off-lattice 336
Configurational-bias Monte Carlo acceptance rule, super-detailed balance 340
Configurational-bias Monte Carlo acceptance rule, trial orientations 341
Conformational-bias Monte Carlo, Recoil growth, versus 374
Consta, S. 375 382
Constrained dynamics, averages 415
Constrained dynamics, case study 427
Constrained dynamics, probability density 41
Constrained dynamics, SHAKE 427
Coordinate transformation, canonical 489
Coulomb potential 292
Cracknell, R.F. 329
Creutz,M. 111 114
Crippen, G.M. 399
Critical exponents 217
Crooks, G.E. 196 198
Crozier,P.S. 318
Csajka,F.S. 450 456
Cui,S.T. 374
Cummings, P.T. 168 374
Darden, T.A. 292 311 312 313 314 316
Davis, H.T. 135 221
De Gennes, P.G. 222
de Leeuw, S.W. 170 222 292 312 317 415
de Miguel, E. 220 223
de Pablo, J.J. 235 271 331 372 374 394 395 397
de Smedt, Ph. 168 216 218 567
de Swaan Arons, J 223
Deem, M.W. 42 358 359 360 386 393
Deitrick, G.L. 221
Dellago, C 450 453 456 462
Deserno,M. 311 312 314
Detailed balance 42 112
Detailed balance, biased configurations 323
Detailed balance, canonical ensemble 114
Detailed balance, case study 54
Detailed balance, grand-canonical ensemble 130
Detailed balance, Metropolis scheme 29
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте



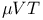 ensemble
ensemble