јвторизаци€
ѕоиск по указател€м
Leach A.R. Ч Molecular Modelling Principles and Applications
ќбсудите книгу на научном форуме Ќашли опечатку?
Ќазвание: Molecular Modelling Principles and Applicationsјвтор: Leach A.R. јннотаци€: Preface to the Second Edition The impetus for this second edition is a desire to include some of the new techniques that have emerged in recent years and also extend the scope of the book to cover certain areas that were under-represented (even neglected) in the first edition. In this second volume there are three topics that fall into the first category (density functional theory, bioinformatics/protein structure analysis and chemoinformatics) and one main area in the second category (modelling of the solid state). In addition, of course, a new edition provides an opportunity to take a critical view of the text and to re-organise and update the material. Thus whilst much remains from the first edition, and this second book follows much the same path through the subject, readers familiar with the first edition will find some changes which I hope they will agree are for the better. As with the first edition we initially consider quantum mechanics, but this is now split into two chapters. Thus Chapter 2 provides an introduction to the ab initio and semi-empirical approaches together with some examples of the uses of quantum mechanics. Chapter 3 covers more advanced aspects of the ab initio approach, density functional theory and the particular problems of the solid state. Molecular mechanics is the subject of Chapter 4 and then in Chapter 5 we consider energy minimisation and other 'static' techniques. Chapters 6, 7 and 8 deal with the two main simulation methods (molecular dynamics and Monte Carlo). Chapter 9 is devoted to the conformational analysis of 'small' molecules but also includes some topics (e.g. cluster analysis, principal components analysis) that are widely used in informatics. In Chapter 10 the problems of protein structure prediction and protein folding are considered; this chapter also contains an introduction to some of the more widely used methods in bioinformatics. In Chapter 11 we draw upon material from the previous chapters in a discussion of free energy calculations, continuum solvent models, and methods for simulating chemical reactions and defects in solids. Finally, Chapter 12 is concerned with modelling and chemoinformatics techniques for discovering and designing new molecules, including database searching, docking, de novo design, quantitative structure-activity relationships and combinatorial library design. As in the first edition, the inexorable pace of change means that what is currently considered 'cutting edge' will soon become routine. The examples are thus chosen primarily because they illuminate the underlying theory rather than because they are the first application of a particular technique or are the most recent available. In a similar vein, it is impossible in a volume such as this to even attempt to cover everything and so there are undoubtedly areas which are under-represented. This is not intended to be a definitive historical account or a review of the current state-of-the-art. Thus, whilst I have tried to include many literature references it is possible that the invention of some technique may appear to be incorrectly attributed or a 'classic' application may be missing. A general guiding principle has been to focus on those techniques that are in widespread use rather than those which are the province of one particular research group. Despite these caveats I hope that the coverage is sufficient to provide a solid introduction to the main areas and also that those readers who are 'experts' will find something new to interest them.
язык: –убрика: ’ими€ /—татус предметного указател€: √отов указатель с номерами страниц ed2k: ed2k stats »здание: 2-nd√од издани€: 2001 оличество страниц: 774ƒобавлена в каталог: 21.02.2007ќперации: ѕоложить на полку |
—копировать ссылку дл€ форума | —копировать ID
ѕредметный указатель
Conformational analysis, clustering algorithms and pattern recognition 491Ч497 Conformational analysis, conformational search 457 662 Conformational analysis, crystal structures predicted 501Ч505 Conformational analysis, dimensionality of data set reduced 497Ч499 Conformational analysis, distance geometry 467Ч475 476 Conformational analysis, fitting, molecular 490Ч491 Conformational analysis, global energy minimum 458 479Ч483 Conformational analysis, model-building 464Ч465 Conformational analysis, poling 499Ч501 Conformational analysis, random 465Ч467 476 Conformational analysis, structural databases 482 489Ч490 493Ч494 499 Conformational analysis, systematic 458Ч464 476 505 Conformational analysis, variations on standard methods 477Ч479 Conformational changes in molecular dynamics simulation 392Ч393 Conformationally flexible docking 662Ч663 CONGEN program 542 Conjugate gradients 262 264Ч267 473 Conjugate peak refinement 290Ч291 Connection table 643 Consistent force field (CFF) 231 Constraints, chiral 473Ч474 Constraints, holonomic versus non-holomonic 370 Constraints, holonomic versus non-holomonic and restraints, difference between 369Ч370 Constraints, holonomic versus non-holomonic in molecular dynamics 368Ч374 Constraints, holonomic versus non-holomonic, simulation 368Ч374 Constraints, holonomic versus non-holomonic, subset selection 717 Constraints, holonomic versus non-holomonic, systematic search 649Ч651 Contact surface 7 Continuum models and solvation, free energy of 592Ч593 598Ч601 Contraction, basis set 69 Convergence sphere 186 coordinates 2Ч4 Coordinates, internal 2Ч4 257 Coordinates, intrinsic reaction 288Ч289 Coordinates, mass-weighted 274Ч275 Coordinates, scaled 438Ч439 (see also УCartesian coordinatesФ) Corey Ч Pauling Ч Koltun (CPK) models 5Ч6 CORINA program 659 Correlation, BLYP 135 136 137 Correlation, coefficients 374 681 Correlation, electron 110Ч117 Correlation, exchange-correlation functional 129Ч134 Correlation, functions and molecular dynamics, simulation 374Ч380 Correlation, spectroscopy (COSY) 474Ч475 486 Cosine coefficient 676 685 COSMO (conductor-like screening model) 597 COSY (correlated spectroscopy) 474Ч475 486 Coulomb interaction/integral 598Ч599 Coulomb interaction/integral, advanced ab initio methods 122 127 128 132 133Ч134 146Ч147 Coulomb interaction/integral, computational quantum mechanics 30 42 45 49Ч53 58 60 85 100 Coulomb interaction/integral, force fields 184Ч185 187 194 202 238 Coulomb potential 167 244 338 341Ч342 CoulombТs law 95 194 202 212 596 603Ч604 607 Counterpoise correction 121Ч122 Coupling parameter 567 Craig plot 681 Cross-correlation function 376 Crossover operator 480Ч481 Crystal momentum 148 Crystal structures, predicted 501Ч505 CSD (Cambridge Structural Database), conformational analysis 482 489 493Ч494 499 CSD (Cambridge Structural Database), new molecules 659 691 693 Cu-Zn superoxide dismutase 607 Cut-offs in computer simulation 324Ч327 Cut-offs in computer simulation, group-based 327Ч330 Cut-offs in computer simulation, problems with 330Ч334 Cyclic urea, HIV protease inhibitor 691Ч693 Cyclobutane 176Ч177 Cyclobutanone 176 Cyclobutene 117 Cycloheptadecane 476 Cyclohexane 286 463 465 497 597 644 Cyclopropene 117 Cyclosporin 196Ч197 391 Cysteine 511 525 556Ч557 Cytosine 84 227 D-Glucose 575 D-optimal design 697Ч698 databases 489 537 539 Databases, 3D 659Ч661 679 УStructural Davidon Ч Fletcher Ч Powell method 269Ч270 de Broglie thermal wavelength 411 440Ч441 de novo ligand design 687Ч694 Degrees of freedom 699Ч700 Delocalised 233Ч234 DelPhi program 604Ч605 Density, charge density matrix 58Ч59 Density, electron 77Ч79 80Ч2 160 Density, functional theory 126Ч137 156 619 Density, functional theory of levels 154 Density, spin 129Ч131 Density, spin of states and Fermi surface 153Ч155 Depth-first search 462 663 Derivative, energy, calculating 120Ч121 Derivative, energy, function 225Ч226 Derivative, energy, minimisation 257Ч258 261Ч262 268Ч269 Descriptors and new molecules 668Ч679 Determinant of matrix 13Ч14 DFP (Davidon Ч Fletcher Ч Powell) method 269Ч270 DFT (density functional theory) 126Ч137 156 619 DHFR (dihydrofolate reductase) 278Ч279 320 460 Diagonalisation of matrix 16 Diatomic overlap see УMNDOФ Dick Ч Overhauser shell model 239 dielectric constant 297 Dielectric models 202Ч204 Diels Ч Alder reaction 294Ч295 615 716Ч717 Differences, free energy 564Ч574 Differences, free energy, applications of methods 569Ч574 Differences, free energy, formula for 568 630Ч631 Differences, free energy, methods for calculating 564Ч569 Differential overlap, neglect of 86 89Ч96 Differential overlap, zero (ZDU) 88Ч89 91 Dihedral angle, definition 4 Dihydrofolate reductase 278Ч279 320 460 DIIS (direct inversion of iterative subspace) 118 Dimensionality reduction 497Ч499 Dimethyl formamide 613Ч614 Dimethyl thioether 676Ч677 Dipole 75Ч77 Dipole correlation time 378Ч378 Dipole, force fields 181 182Ч185 189 199Ч201 219 246 Dipole, models of solvation, free energy of 593Ч595 601Ч603 Dipole, moment, net 378Ч379 Dipole, triple 213Ч215 239 Direct inversion of iterative subspace 118 Direct SCF method 118Ч120 Director 396 Discriminant analysis and QSAR 703Ч705 Dispersion curve 298Ч299 Dispersive interactions 204Ч206 Displacements 322Ч323 624 Dissimilarity-based methods 683Ч685 Dissipative particle dynamics 402Ч404 Distance, bounds 468 Distance, bounds, geometry 467Ч475 476 651Ч653 663 Distance, bounds, Hamming (city block) 492 676Ч678 Distance, bounds, map 651 Distance, bounds, matrix 652 Distance, bounds, Soergel 676Ч678 Distance-dependent dielectric 203 Distributed multipole analysis 195Ч197 Diverse sets of compounds, selecting 680Ч687 DMA (distributed multipole analysis) 195Ч197 DMF (dimethyl formamide) 613Ч614 DNA 197 227 452 489 604 DNA, computer simulation 319 338Ч339 DNA, Human Genome Project 512 548Ч549 DNA, inhibitor 270Ч271 DNA, new molecules 662 DNA, proteins 509 512 549 DOCK program/algorithm 662Ч663 665 667 docking 661Ч668 689 Domain 515 Double dynamic programming 537Ч539 Double zeta basis sets 70 Double-wide sampling 567Ч568 DPD (dissipative article dynamics) 402Ч404 Dreiding models 5Ч6 Drude molecules 205Ч206 Drude molecules, interaction between 246Ч247 Drugs, new see УNew moleculesФ Dual topology 578 Dummy atoms, in Z-matrix 271Ч272 Dunning basis sets 73 Dynamic/ dynamics 353Ч409 Dynamically modified windows 578 Dynamically modified windows and statics in energy minimisation 295Ч300 (see also УMolecular dynamicsФ) Dynamically modified windows, programming and protein prediction 526Ч529 EA (evolutionary algorithms) 479Ч483 Edges of search trees 461 Effective medium theory 243Ч241 Effective pair potentials 214Ч215 Eigenvalues and eigenvectors 15Ч16 114 Eigenvalues and eigenvectors, conformational analysis 469 471 479 498 Eigenvalues and eigenvectors, energy minimisation methods 272 282Ч285 Einstein relationships 381 627Ч628 EisenbergТs 3D profiles 543Ч544 Elastic constants 240 296Ч297 Electric multrpoles, calculation of 75Ч77 Electron, affinity (EA) 194 Electron, correlation 110Ч117 Electron, density 77Ч79 80Ч82 160 Electron, gas theory 240 Electron, integrals, one- and two- 50Ч51 Electron, nearly free-electron approximation 147Ч153 Electron, polyelectronic atoms and molecules 34Ч41 Electron, spin 34Ч35 38 Electronegativity 192Ч193 Electrostatic interactions, force fields 166 181Ч204 221 237 Electrostatic interactions, free energy calculations 566Ч567 576 580 588 613 Electrostatic interactions, potentials 83Ч85 188 189Ч191 Electrostatic interactions, solvation free energy calculations 593Ч608 Electrotopological state index 674 Embedded-atom model 241 243Ч244 Embedding 469 Empirical bond-order potential see УTersoffФ Endothiapepsin 589Ч591 Energy minimisation methods 253Ч302 623 Energy minimisation methods, applications of 273Ч279 Energy minimisation methods, choice of 270Ч273 Energy minimisation methods, derivative 257Ч258 261Ч262 268Ч269 Energy minimisation methods, first-order 262Ч267 Energy minimisation methods, Newton Ч Raphson 267Ч268 270 288 Energy minimisation methods, non-derivative 258Ч261 Energy minimisation methods, quasi-Newton 268Ч269 Energy minimisation methods, solid-state systems 295Ч300 Energy minimisation methods, statement of problem 255Ч257 Energy minimisation methods, transition structures and reaction pathways 279Ч295 Energy of closed-shell system 51 Energy of general polyelectronic system 46Ч50 Energy, calculation from wavefunction 41Ч46 Energy, component analysis 122Ч124 Energy, computer simulation 308 348Ч349 Energy, conservation in molecular dynamics simulation 359 405Ч406 Energy, derivatives, calculating 120Ч121 Energy, force field 240 Energy, function, derivatives of 225Ч226 Energy, global minimum 253 458 479Ч483 551Ч552 Energy, KoopmanТs theorem and iorusation potentials 74Ч75 Energy, lower-energy regions 564 Energy, minimum, global 253 458 479Ч483 551Ч552 Energy, potential 4Ч5 238 253 Energy, strain 226Ч7 627 Energy, surface (hypersurface) 4Ч5 253 475 Energy, units of 9 (see also УDerivativeФ УEnergy УFree УQuantum Enol boroate/aldehyde reaction 610Ч611 Ensemble, averages 303Ч305 Ensemble, distance geometry 651Ч653 Ensemble, molecular dynamics 653 Ensemble, Monte Carlo simulation 438Ч442 450Ч451 Enthalpy 159 574 entropy 574 Enumeration of libraries 715Ч717 Equilibration monitoring and computer simulation 321Ч323 Equilibria phases in computer simulation 315 450Ч451 Ergodicity 304 313 Ergodicity, quasi ergodicity 433Ч438 Error, estimating in a simulation 343Ч347 ESS (explained sum of squares) 699Ч700 Ethane, carbon-carbon bond 4Ч5 Ethane, force fields 174 Ethane, Monte Carlo simulation 441Ч442 Ethane, SMILES notation 644 Ethane, thiol 564Ч569 Ethane, torsion angles 286Ч287 Ethane, Z-matrix 2Ч3 9 Ethanol 564Ч569 611Ч612 Ethene 83 236 293Ч295 ethylene 621 Ethyne 83 Euclidean distance measure 492 Euler angles 421Ч422 Even-tempered basis set 71Ч72 Evolutionary algorithms 479Ч483 Evolutionary design 694 Evolutionary planning (EP) and strategies (ES) 479Ч480 482 Ewald summation 238 334Ч339 342 402 625Ч626 Exchange, correlation functional 129Ч134 Exchange, forces see УRepulsive forcesФ Exchange, gradient 135 136 137 Exchange, integral 50 52Ч53 58 60 Exchange, interaction 46 Exclusion spheres 658Ч659 Exons 512 Explained sum of squares 699Ч700 Extended Hiickel theory (EHT) 101Ч102 Extended system method 384 Extreme value distribution 532 Fabric softeners 401Ч402 Face-centred cubic lattice 139 316 Factor analysis 681Ч682 686 Factors (variables) 697 Family (proteins) 539 Fast Fourier Transform 24 338Ч339 342 Fast multipole method 341Ч343 364 FASTA 524 531 FDPB (finite difference Poisson Ч Boltzmann method) 604Ч608 Fermi surface and energy 153Ч155 Ferrocene 234 FFT (fast Fourier transform) 24 338Ч339 342 FickТs laws 380Ч381 Finite difference methods 355Ч358 604Ч608 Finnis Ч Sinclair potential 241Ч245 passim First principles method for predicting proteins 517Ч522 First-order energy minimisation 262Ч267 Fitting, molecular 490Ч491 Flexible fitting 491 Flexible molecules 423 582Ч585 FlexX program 667 Fock matrix 100 Fock matrix, Hartree Ч Fock equations 57Ч59 61 63Ч64 Fock matrix, open-shell systems 108Ч109 Fock matrix, semi-empirical methods 89Ч90 94 95 96 Fock matrix, solid state quantum mechanics 146 Fock operator 53 57 114 Focusing 606 Folding see УUnder proteinsФ Force field models, empirical 165Ч252 610 Force field models, empirical, 233Ч234
–еклама
 |
|
ќ проекте
|
|
ќ проекте



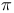 -systems, force fields for
-systems, force fields for