јвторизаци€
ѕоиск по указател€м
Leach A.R. Ч Molecular Modelling Principles and Applications
ќбсудите книгу на научном форуме Ќашли опечатку?
Ќазвание: Molecular Modelling Principles and Applicationsјвтор: Leach A.R. јннотаци€: Preface to the Second Edition The impetus for this second edition is a desire to include some of the new techniques that have emerged in recent years and also extend the scope of the book to cover certain areas that were under-represented (even neglected) in the first edition. In this second volume there are three topics that fall into the first category (density functional theory, bioinformatics/protein structure analysis and chemoinformatics) and one main area in the second category (modelling of the solid state). In addition, of course, a new edition provides an opportunity to take a critical view of the text and to re-organise and update the material. Thus whilst much remains from the first edition, and this second book follows much the same path through the subject, readers familiar with the first edition will find some changes which I hope they will agree are for the better. As with the first edition we initially consider quantum mechanics, but this is now split into two chapters. Thus Chapter 2 provides an introduction to the ab initio and semi-empirical approaches together with some examples of the uses of quantum mechanics. Chapter 3 covers more advanced aspects of the ab initio approach, density functional theory and the particular problems of the solid state. Molecular mechanics is the subject of Chapter 4 and then in Chapter 5 we consider energy minimisation and other 'static' techniques. Chapters 6, 7 and 8 deal with the two main simulation methods (molecular dynamics and Monte Carlo). Chapter 9 is devoted to the conformational analysis of 'small' molecules but also includes some topics (e.g. cluster analysis, principal components analysis) that are widely used in informatics. In Chapter 10 the problems of protein structure prediction and protein folding are considered; this chapter also contains an introduction to some of the more widely used methods in bioinformatics. In Chapter 11 we draw upon material from the previous chapters in a discussion of free energy calculations, continuum solvent models, and methods for simulating chemical reactions and defects in solids. Finally, Chapter 12 is concerned with modelling and chemoinformatics techniques for discovering and designing new molecules, including database searching, docking, de novo design, quantitative structure-activity relationships and combinatorial library design. As in the first edition, the inexorable pace of change means that what is currently considered 'cutting edge' will soon become routine. The examples are thus chosen primarily because they illuminate the underlying theory rather than because they are the first application of a particular technique or are the most recent available. In a similar vein, it is impossible in a volume such as this to even attempt to cover everything and so there are undoubtedly areas which are under-represented. This is not intended to be a definitive historical account or a review of the current state-of-the-art. Thus, whilst I have tried to include many literature references it is possible that the invention of some technique may appear to be incorrectly attributed or a 'classic' application may be missing. A general guiding principle has been to focus on those techniques that are in widespread use rather than those which are the province of one particular research group. Despite these caveats I hope that the coverage is sufficient to provide a solid introduction to the main areas and also that those readers who are 'experts' will find something new to interest them.
язык: –убрика: ’ими€ /—татус предметного указател€: √отов указатель с номерами страниц ed2k: ed2k stats »здание: 2-nd√од издани€: 2001 оличество страниц: 774ƒобавлена в каталог: 21.02.2007ќперации: ѕоложить на полку |
—копировать ссылку дл€ форума | —копировать ID
ѕредметный указатель
701 699 612Ч614 280Ч282 513Ч515 513Ч514 513 197Ч199 126 233Ч234 1, 2-dichloroethane 387Ч388 1, 3, 5-trifluorobenzene 198Ч199 1-octanol and water, partition between 668Ч669 2-methyl propane 644 2D substructure searching 642Ч647 4-acetamido benzoic acid 661 4-methyl-2 oxetanone 137 ab initio defined 65 ab initio molecular dynamics 616Ч622 ab initio potentials for water 216Ч218 ab initio quantum mechanics, calculating properties using 74Ч86 (see also УAdvanced ab initio methodsФ) Absolute free energies 573Ч574 Accessible surface 7 ACE (angiotensin converting enzyme) 649Ч651 Acetaldehyde 180 578 Acetamide 573 Acetic acid 504Ч505 573 643 Acetic acid, SMILES notation 644 645 Acetonitrile 597 Acetylcholine 678 Acronyms and abbreviations 104Ч105 553Ч554 Adenine 227 Adiabatic mapping 286 Adjacency matrix 647 Adjoint matrix 15 Adsorption processes, Monte Carlo simulations of 441Ч442 Advanced ab initio methods 108Ч164 Advanced ab initio methods, density functional theory 126Ч137 Advanced ab initio methods, electron correlation 110Ч117 Advanced ab initio methods, energy component analysis 122Ч124 Advanced ab initio methods, open-shell systems 108Ч110 Advanced ab initio methods, practical considerations 117Ч122 Advanced ab initio methods, solid state quantum mechanics 138Ч160 Advanced ab initio methods, valence bond theories 124Ч126 Agglomerative cluster analysis methods 493Ч494 Agonists 640 AINT function 350 Alanines 169 459 511 525 542 546 556Ч557 Alanines, energy minimisation methods 277 280 286 Alanines, free energy calculations 583Ч584 Aldehyde 610Ч611 Aldol reactions 610Ч612 Aliovalent substitution 623 Alkaline earth oxides 147 Alkanes 449Ч450 AMBER force field 169Ч170 175Ч176 191 211 230Ч232 AMI 86 97Ч98 102Ч103 230 Amino acids 511 549 602 Amino acids, computer simulation 329Ч330 Amino acids, conformational analysis 459 487 Amino acids, energy minimisation methods 277 280 286 Amino acids, force fields 169Ч170 221 Amino acids, free energy calculations 572 583Ч584 Amino acids, motifs 522 Amino acids, PAM matrices 524Ч526 531 556Ч557 Amino acids, torsion angles 515 (see also УPeptides; proteinsФ) Amino acids, ТthreadingТ 546 Aminothlazoles 716 AMP AC program 8 99 Amphiphiles, molecular dynamics simulation of 394Ч404 Angiotension converting enzyme 649Ч651 Angle bending 166 173 Annealing, simulated 483Ч489 504Ч505 519 691 Annotations 513 Antagonists 640 Antisymmetry principle 35 Arbitrary step energy minimisation 264 Argand diagram 17 Arginme 329Ч330 510 525 556Ч557 Argon 253 323 Argon, force fields 205 214 Argon, J-walking 434Ч435 Argon, time-steps 361Ч362 Argon, velocity autocorrelation 377 Arithmetic mean 20 Aromatic systems and charge schemes 197Ч199 Asparagine 330 510 525 546 556Ч557 Aspartic acid 510 525 556Ч557 Atoms in molecules theory 80Ч81 Atoms/atomic, charges 157Ч159 181 192Ч195 Atoms/atomic, marker 329Ч330 Atoms/atomic, one-electron 30Ч34 Atoms/atomic, orbitals 41Ч42 56 100 241 Atoms/atomic, polyelectronic 34Ч41 Atoms/atomic, type 169 Atoms/atomic, units 29 Aufbau principle 35 Austin Model I (AMI) 86 97Ч98 102Ч103 Autocorrelation function 376Ч378 Automated protein modelling 548Ч549 Autoscaling 681 Availability 300 Axilrod Ч Teller term (triple-dipole) 213Ч215 239 Azimuthal quantum number 31 B3LYP density function 615 Backtracking 462 Backward sampling 567 band theory 141Ч142 Band theory, orbital-based approach 142Ч146 Barker Ч Fisher Ч Watts potential 214 Basic Local Alignment Search Tool see УBLASTФ Basic Local Alignment Search Tool, basis sets/functions 56 85 123 Basic Local Alignment Search Tool, computational quantum mechanics 65Ч74 Basic Local Alignment Search Tool, Gaussian functions 65Ч73 passim 120 137 195 Basic Local Alignment Search Tool, superposition error 121Ч122 (see also УSTOsФ) BCUT method 686Ч687 Bead model of polymers 428 Becke see УBLYPФ BeemanТs algorithm 357 Bending 166 173 176Ч178 Benperidol 675 Benzamidine 586Ч588 benzene 123 Benzene, force fields 170 174 178 186 197Ч198 Benzene, Huckel theory 99 100 Benzene, ring 81 170 178 Benzene, SMILES notation 644 Benzene, spin-coupled valence bond theory 126 Benzyl bromide 670 Beryllium 40 113 BFGS(Broyden-Fletcher-Goldfarb-Shanno) method 269Ч270 Bilinear model 702 Binding site 662 689Ч691 Binomial expansion 11 Bioinformatics 513 (see also УAmino acidsФ УDNAФ УProteinsФ) Bioisosteres 648 Biotin 576 641 bitstring 645Ч647 BLAST (Basic Local Alignment Search Tool) 521 524 531Ч534 548 BlochТs theorem/function 142Ч145 146 148 161 Block-diagonal Newton Ч Raphson minimisation 268 BLOSUM matrices 526 BLYP (Becke gradient-exchange correction and Lee Ч Yang Ч Parr correlation functional) 135 136 137 Bohr radius 31 Boltzmann distribution 192 214 274 483 611 Boltzmann distribution, computer simulation 306Ч307 347 Boltzmann distribution, conformational analysis 457 Boltzmann distribution, Monte Carlo simulation 415Ч416 433 435Ч436 445Ч446 Boltzmann factor 306Ч307 413 Boltzmann weighted average 576 581 Bond fluctuation model 424Ч425 Bond/bonding 644 Bond/bonding, inorganic molecules 234Ч235 Bond/bonding, lack of see УNon-bonded interactionsФ Bond/bonding, orders 81Ч83 Bond/bonding, stretching 166 170Ч173 Bond/bonding, valence 124Ч126 (see also УUnder carbonФ УHydrogenФ) Born equation/model 238 593Ч594 598Ч601 609 Born Ч Oppenheimer approximation 4 35Ч36 50 Boundary, computer simulation 317Ч321 Boundary, element method 598 Bravais lattices 138Ч139 BrillouinТs theorem 112Ч113 115 Brilloum zone 140Ч141 145Ч146 150Ч151 157Ч158 298Ч299 Bromine 571 Broyden Ч Fletcher Ч Goldfarb Ч Shanno method 269Ч270 BSSE (basis set superposition error) 121Ч122 Buckingham potential 209 238 Build-up approach 517 Butadiene 233 294Ч295 Butane 85Ч86 186 449Ч450 582 Butanone 611Ч612 Cage structure of liquids 377 Calcium 589 626 Calix[4]arene 291Ч292 Cambridge structural database see УCSDФ canonical ensemble 563 569 Canonical genetic algorithms 479Ч480 Canonical representation 644 Canonical structures 541 Captopril 649 Car Ч Parrinello scheme 610 617Ч619 Carbo index 678Ч679 Carbon dioxide 181 316 616Ч617 Carbon, bonds 4Ч5 98 362 612 652 Carbon, bonds, energy minimisation methods 253 280Ч281 Carbon, bonds, force fields 167 180 211 233 236 Carbon, five-carbon fragment 472 Carbon, force fields 167 180 211 233 236 244 Carbon, valence electron density 160 Carbonic anhydrase 616 Carboxylic acid 660 Cartesian coordinates 2Ч4 Cartesian coordinates, conformational analysis 466Ч468 Cartesian coordinates, energy minimisation methods 255 257Ч258 275 290 Cartesian coordinates, molecular dynamics simulation 370 372 379 393 Cartesian coordinates, Monte Carlo simulation 417 420Ч421 423 Cartesian coordinates, vectors 11Ч12 CASP3 548 CASSCF (complete active-space SCF) 113 295 CAVEAT program 689 CBMC (configurational bias Monte Carlo) simulation 443Ч50 Cell, cubic 315Ч319 Cell, index method 326 Cell, multipole method 341Ч343 Cell, unit 138 Cell, Wigner Ч Seitz 140 350 Central dogma 509 512 Central multipole expansion 181Ч187 CFF (consistent force field) 231 Chain amphiphiles and molecular dynamics 394Ч404 Charge 187 Charge, atomic 157Ч159 181 191Ч195 Charge, density matrix 58Ч59 Charge, image simulation 340Ч341 Charge, oscillating 201Ч202 Charge, schemes 197Ч199 CHELP procedure 191 Chemical potential calculation in Monte Carlo simulation 442Ч443 Chemical reactions 610Ч622 Chemical reactions, molecular dynamics 616Ч622 Chemical reactions, potential of mean force of 612Ч614 Chemical reactions, quantum and molecular mechanics combined 614Ч616 Chemical reactions, simulation empirically 610Ч612 Chemokine 475 Chi molecular connectivity indices 672 Chi-squared test 344Ч345 Chiral constraints 473Ч474 Chlorides 238 612Ч613 614 676Ч677 Chlorine 181 231 280Ч281 571 612 620Ч621 Chloroform 227 573 Chlorpromazine 678 Chymosin 545 Chymotrypsin 522Ч523 CI (configuration interaction) 111Ч113 120 CID (configuration interaction doubles) 112 113 CISD (configuration interaction singles and doubles) 112 113 City block see УHammingФ Clausius Ч Mosotti relationship 238Ч239 Clausius, virial theorem of 309 Clique detection 653Ч656 CLOGP program 669Ч670 Closed-shell systems 51 56Ч9 86Ч88 109 Cluster analysis 494 534 682Ч683 Clustering algorithms 491Ч497 CMR program 671 CNDO (complete neglect of differential overlap) 86 89Ч92 93 94 95 Coefficients 148Ч149 374 Coefficients, new molecules 676Ч678 680Ч681 685 Coefficients, partition 572Ч573 668Ч671 Cofactor 14Ч15 Combination generator 420 453Ч454 Combinatorial explosion 460Ч461 Combinatorial libraries 711Ч719 CoMFA (comparative molecular field analysis) 679 708Ч711 Comparative modelling of proteins 539Ч545 Comparative molecular field analysis 679 708Ч711 Complete active-space SCF 113 295 Complete neglect of differential overlap 86 89Ч92 93 94 95 Complete-linkage (furthest-neighbour) cluster algorithm 493Ч494 complex numbers 16Ч18 Computational quantum mechanics 26Ч107 Computational quantum mechanics, acronyms used in 104Ч105 Computational quantum mechanics, approximate orbital theories 86 Computational quantum mechanics, atomic units 29 Computational quantum mechanics, basis sets 65Ч74 Computational quantum mechanics, calculating properties 74Ч86 Computational quantum mechanics, Huckel theory 99Ч102 Computational quantum mechanics, one-electron atoms 30Ч34 Computational quantum mechanics, operators 28Ч29 Computational quantum mechanics, polyelectroruc atoms and molecules 34Ч41 Computational quantum mechanics, semi-empirical methods 65 86Ч99 102Ч103 УHartree Computer simulation 303Ч352 Computer simulation, boundaries 317Ч321 Computer simulation, equilibration, monitoring 321Ч323 Computer simulation, free energy calculation difficulties 563Ч564 Computer simulation, long-range forces 334Ч343 Computer simulation, molecular dynamics 305Ч306 307 Computer simulation, phase space 312Ч315 Computer simulation, practical aspects 315Ч316 Computer simulation, real gas contribution to virial 309 349Ч350 Computer simulation, results and errors 343Ч347 Computer simulation, statistical mechanics 347Ч348 Computer simulation, thermodynamic properties, simple 307Ч312 Computer simulation, time and ensemble averages 303Ч305 Computer simulation, translating particle back into control box 350 Computer simulation, truncating potential and minimum image convention 324Ч334 (see also УMolecular dynamics simulation Monte Computers, hardware 8Ч9 Computers, Internet and World Wide Web 9Ч10 548 553 Computers, software 8Ч9 99 concepts 1Ч25 (see also УCoordinatesФ УMathematical CONCORD program 659 Conditionally convergent series 336 Conduction band 142 Conductor-like screening model 597 Configuration interaction see УCIФ УCIDФ УCISDФ Configurational bias see УCBMCФ Conformational analysis 457Ч508 Conformational analysis and NMR/x-ray crystallography 468 474Ч475 483Ч489 Conformational analysis, choice of method 476Ч477
–еклама
 |
|
ќ проекте
|
|
ќ проекте



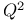 (cross-validated R2)
(cross-validated R2) 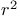
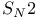 reaction, potential of mean force
reaction, potential of mean force  -Helix
-Helix  -strand structures
-strand structures  systems
systems