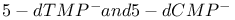|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Leontis N.B. (ed.), SantaLucia J., Jr. (ed.) — Molecular Modeling Of Nucleic Acids |
|
|
 |
| Предметный указатель |
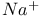 counterions, influence on gas-phase ionization potentials 30—34 counterions, influence on gas-phase ionization potentials 30—34
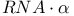 -winged ODN hybrids, denaturing gel electrophoretic analysis of RNase H digestion products 95f -winged ODN hybrids, denaturing gel electrophoretic analysis of RNase H digestion products 95f
 -anomeric nucleotides, base stacking and specific base pairing 103 -anomeric nucleotides, base stacking and specific base pairing 103
 -containing decamer duplexes, -containing decamer duplexes,  P NMR spectra 104 P NMR spectra 104
 -containing decamer duplexes, imino proton region 98 3 -containing decamer duplexes, imino proton region 98 3
 -containing decamer duplexes, sequences 96 -containing decamer duplexes, sequences 96
 -containing decamer duplexes, sugar puckering data 100 -containing decamer duplexes, sugar puckering data 100
 -containing decamer duplexes, thermodynamic data 971 -containing decamer duplexes, thermodynamic data 971
 -containing DNA duplexes, spectroscopic studies 94 103 -containing DNA duplexes, spectroscopic studies 94 103
 -winged ODN, RNA targets 94 -winged ODN, RNA targets 94
 -winged ODN/RNA duplexes, cleavage by RNase H 92 94 -winged ODN/RNA duplexes, cleavage by RNase H 92 94
 -winged RNase H substrates, sequences and control 95 -winged RNase H substrates, sequences and control 95
 A decamer duplexes, models 102 A decamer duplexes, models 102
 C decamer, pathways 99 C decamer, pathways 99
 T decamer, studies 103 T decamer, studies 103
 stretch, schematic diagram 93 stretch, schematic diagram 93
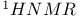 spectra of spectra of  imino proton and methyl group regions 104 imino proton and methyl group regions 104
 , backbone conformation of duplexes 100 , backbone conformation of duplexes 100
3-Hydroxytetrahydrofuran, photoelectron spectra, molecular orbital diagrams, and IPs 20f
3D NOE-NOE refinements, quality 173
3D NOESY-NOESY refinement, methods 174—176
3D NOESY-NOESY refinement, simulation study 170
9-Methyladenine, photoelectron spectra, molecular orbital diagrams, and IPs 21f
A-DNA, stabilized by hydration and counterion association in major groove 299
A-DNA, structure stabilization 297
A-form geometry, stability 296
A-tract DNA, interpretation of Raman bands 152 155
A-tract DNA, measurement and assignments of Raman bands 151 152
A-tract DNA, solution structure away from crystal packing constraints 164
A-tract-T-tract (bent) DNA, molecular modeling 155 159
ab initio methods, quantum mechanical, calculations 7 8
Acid-base titration, native DNA 2
Adenine H2 protons, partial relaxation correction 137
Adenosine platforms, same strand pairing 74
Adsorption density, probe sites 213
Adsorption density, saturating match and mismatch 226
Adsorption equilibrium, probe sites 213 217
Adsorption isotherms, matched probes 214
Adsorption kinetics, change with time for matched probes 212
Adsorption kinetics, duplex formation 211 213
Adsorption to probes, matched and single-base mismatched 214
Affymetrix 206
Algorithms, genetic 229 245
AMBER, all-hydrogen parameter sets 304
AMBER, counterion equilibration 174
AMBER, distrance constraints 175
AMBER, electrostatic potential energy 319
AMBER, energy functions 262
AMBER, energy minimization 113
AMBER, nab molecular mechanics package 386
AMBER, refinement calculations 140
AMBER, sodiums within contact distance with phosphates 333
AMBER, spontaneous transitions between A and B forms of DNA 305
AMBER3.0, force field results 266 270
AMBER4.0, ring-saturated thymine derivatives 318
AMBER4.1, conformational behavior of oligonucleotides in aqueous solution 289
AMBER4.1, DNA conformational transitions 278
AMBER4.1, dynamical models of oligonucleotide sequences 274
AMBER4.1, model compound calculations 45
AMBER4.1, model compounds for parameterization 44
AMBER4.1, molecular simulations 48—49
AMBER4.1, simulating proteins and nucleic acids 43
AMBER4.1, simulation of mlp 331
Amino acid, sequence SPXX in nuclear proteins 195 204
Andre, Troy C. 246
Anisotropic duplexes, cross-relaxation rate, constants 111
Anisotropic motions, nucleic acids 110 111
Antiparallel P-sheet motif, binds to minor, groove 196
Antisense DNA therapy, gene expression 92
Antisense drugs, phosphorothioates 42 43
Antisense drugs, properties 92
Antisense oligonucleotides, design and characterization 41 42
Antisense oligonucleotides, nuclease-resistant a-anomeric segments 103
ApT pocket, minor groove 270
ApT pocket, structural detail of 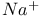 ion 272 ion 272
Aqueous bulk solvation, nucleotide Gibbs, free energies of activation 34 37
Aramini, James M. 92
Artificial nuclease, activity 163
Assembly rules, RNA mosaic units 350 355
Atom truncation, MD simulations of nucleic, acids 304
Atomic resolution models, ribosomal decoding site 372 374
Atomic-level models, nucleic acid, structures 379
Axis bending, DNA sequences 270
Axis bending, spontaneous 274
B — RNA, simulation 288
B-DNA, crystal structures 263—265
B-DNA, marker band 154
B-form trajectory, hydroxyl groups 51
B-form trajectory, protrusion of 2
B-form trajectory, sugar puckering 53
B/Z junctions, double 162
B/Z junctions, location in T-tract-CG-tract-A-tract deoxy oligonucleotides 159—163
B/Z junctions, nuclease attack 151
Backbone conformation, influence on A to B equilibrium 307 310
Backbone torsion angles, molecular dynamics 140
Backbone torsion angles, restraints 124
Backbone torsion angles, variable in polynucleotides 4
Backbone, changes in nucleotide conformations 118
Backbone, degrees of freedom 115
Backbone-sugar torsions, helicoidal, parameters 381
Backfolding, nuclease hypersensitive sites 412
Banerjee, A. R. 360
Base inclination energy, A and B forms of DNA 309
Base locations, construction 381
Base orbitals, comparisons 28
Base orbitals, molecular orbital diagrams 29
Base pair step parameters 3411 3421
Base pair step(s), conformation 337 338
Base pair(s), constraining a nucleotide 253
Base pair(s), duplex oligonucleotides, hydrogen bonding patterns 77—90
Base pair(s), isolated 253
Base pair(s), percent correctly predicted 254
Base pair(s), phylogenetically determined 246
Base pair(s), scoring against phylogenetic structures 253
Base pair(s), terminal fraying 291
Base pairing, algorithms and computer programs 3
Base pairing, DNA and RNA 2
Base pairing, severe weakening 319
Base sequence, local DNA structure 2—3
Base stacking, ab initio study 8
Base stacking, RNA 352
Base-first strategies, nucleic acid modeling 381
Belief and plausibility measures, structural hypothesis 402
Bendability, DNA 330 343
Bending angles, repair of photodamaged DNA 293
Bending Dials, bending in structures 261
Bending, DNA bound by TBP 339
Berzal — Herranz, A. 360
Beveridge, D. L. 260
Bifurcated hydrogen bonds, Raman measurements of carbonyl vibrations 159
Billed, Todd M. 122
Binding affinity, chirality of phos-phorothioate center 43
Binding sites, ribozyme 72
Biograf, DNA models 150
Biograf, energy minimization 155—159
Biological evolution, RNA molecules 347
Biological RNA molecules, internal loops 57
Biomolecule, nuclear spins 10
Biphasic kinetics, cleavage and ligation kinetics 364
Biphasic kinetics, ribozyme 360
BMS, parameter set 304
Bond, J. 360
Bounds object, distance geometry 385
Bowling Green State University 1 167
Bridged structures, length dependence 217
Bridging interactions, target concentrations 226
Brown, Tom 77
Bulk hydration, energetic ordering of ionization events 35—37
Bulk hydration, ionization energies of nucleotide clusters 35
Burke, J. M. 360
| Butcher, S. 360
C++ folding algorithm 248
C++ RNA secondary structure prediction 246
Carbonyl environment, bent and straight DNA 159
Carcinogenicity, structure of photodamaged DNA oligomers 291
Case, David A. 379
Catalytic activity, folded structure of RNA 361
Cellulaire du Centre National de la Recherche Scientifique 346
Chain growth, rate of energy improvement 237—238
Chain length, A-tract — T-tract DNA 156
CHARMM 23
CHARMM, all-hydrogen parameter sets 304
CHARMM, consecutive phosphorus-phosphorus distances 334 335
CHARMM, energy functions 262
CHARMM, force fields with balanced solute-solvent and solvent-solvent interactions 286
CHARMM, periodic boundary conditions for hexagonal prism 331
CHARMM, spontaneous transitions between A and B forms of DNA 305
CHARMM, structure on sodium-sodium rdf 333
CHARMM, transitions observed in DNA helices 294
Cheatham, T. E., Ill 285
Chemical changes to DNA, test case for modeling 289—291
Chemical crosslinking, tRNA-mRNA complex 373
Chemical damage, modified base 86
Chemical shift, changes in peptide-DNA hairpin samples 202 203
Chemical shift, database 118
Chemical shift, peptide — DNA sample 199
Chirality, A- and B-form geometries 41—54
Cieplak, P. 285
Cleavage activity, prevention 363
Cleavage kinetics, chase step 364
Cleavage reactions, single-turnover conditions 367
Cleavage, fluorescence energy transfer signal 364
Clover-leaf model, tRNA 5 6
Clusters, 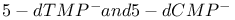 32 32
Clusters, optimization calculations 31
Clusters, structures of 5
Coaxial stacking, helixes 250 363
Coaxial stacking, three-way junction 366
Coaxial stacking, two helixes with intervening mismatch 252
Coaxially oriented helices, SRP RNA 407 410
Code statistics, nab 383
Codon-anticodon interactions, degeneracy of genetic code 80
Cognate DNA sequence, propensity to distort 278
CombiChem, Inc 285
Comparative modeling of 3
Comparative sequence analysis, deducing secondary structures from primary sequence 5—6
Comparative sequence analysis, SRP RNA secondary structures 406
Compensatory base changes, mutations 406
Competing equilibria, probes and target binding 223—224
Computer calculations, quantum mechanical 8
Computer experiment, MD simulation 260—263
Computer folded sequences, approximating free energy parameters of structural motifs 247
Computer language, MC-SYM 380
Computer language, nab 379—393
Computer language, OCL 381
Computer language, X-PLOR 380
Computer language, yammp 380
Computer modeling, characteristics 394—395
Computer modeling, genetic algorithm 229—245
Computer modeling, three-dimensional RNA structures 394—404
Computer software, AMBER 113 140—146 174 175 262 304—311 386
Computer software, AMBER3.0 266—270
Computer software, AMBER4.1 43 45 48 9 274 278 289 331
Computer software, Bending Dials 261
Computer software, BIOGRAF 150 155—159
Computer software, BMS 304
Computer software, C 246 248
Computer software, CHARMM 262 304—311
Computer software, CHARMM23 286 294 331
Computer software, CORMA 184 185—186 189
Computer software, CURVES 68 261 318 331
Computer software, D 405—413
Computer software, Dials and Windows 261 307 318 331
Computer software, DNAminiCarlo 184 185 186 189
Computer software, efn 2 250—252
Computer software, ERNA 3
Computer software, FELIX 174
Computer software, GAUSSIAN 90 314
Computer software, GROMOS 262 265—266
Computer software, Insight II 113 331
Computer software, JUMNA 381
Computer software, MARDIGRAS 124—146 184—186 189
Computer software, MC-SYM 373 395
Computer software, MEDUSA 182 189
Computer software, Metropolis Monte Carlo 184
Computer software, Mfold 246 248
Computer software, mlp 331
Computer software, Molecular Dynamics Tool Chest 261
Computer software, MORASS 174
Computer software, MORCAD 381
Computer software, NEW HELIX 93 156 159
Computer software, Newhelix 261
Computer software, OPLS 286
Computer software, PARSE 182—183 189
Computer software, PDQPRO 185 186 193
Computer software, QUANTA 331
Computer software, RANDMARDI 184
Computer software, SHAKE 290 331
Computer software, Simulaid 331
Computer software, X-PLOR 174 386
Configuration, mRNA-tRNA complexes 372—373
Conformation build-up procedures, nucleic acid modeling 380—381
Conformation, nucleic acids in solution 118
Conformational change, RNA tetraloop 294
Conformational degrees of freedom, duplex DNA 312
Conformational dynamics, DNA backbone torsions 261
Conformational dynamics, spontaneous A-DNA to B-DNA transitions 287
Conformational flexibility, biological, functioning 11
Conformational hell, escape 361—363
Conformational junctions, nonstraight DNA 159—163
Conformational pool, potential conformers for DNA dinucleotide 186
Conformational preferences, calculated and X-ray crystallographic data 315
Conformational preferences, ring-saturated thymine derivatives 313 318
Conformational sampling, problem in simulations 298
Conformational sampling, results of conformer ensemble refinement 189—193
Conformational sampling, structural ensembles 181—194
Conformational sampling, structure of nucleic acids in solution 299—300
Conformational search space, molecular contacts 396—397
Conformational space, base pair step parameters for DNA oligomers 341
Conformational space, RNA 395
Conformational transition, barriers 288
Conformers, generation and selection 183
Conformers, potential 182—183 (see also Potential conformers)
Constraints, factors affecting number and accuracy 108—118
Constraints, structure determination 108
Controlled flexibility, nucleic acids 380
Cooney, Michael 312
CORMA, dipole-dipole relaxation rates 186
CORMA, NOE intensities for each ensemble 185 186 189
CORMA, relaxation rates and NOE intensities 184
CORMA, relaxation rates for potential conformers 185
Cornell et al. force field, conformational behavior of oligo-nucleotides in aqueous solution 289
Cornell et al. force field, DNA conformational transitions 278
Cornell et al. force field, generality 291
Cornell et al. force field, MD simulations on nucleic acid systems 285—303
Correlation function, nucleic acids 111
COSY crosspeaks, estimates of coupling constants 108
Counterion association, A-DNA, stabilization 299
Counterion condensation, MD results and theory predictions 270
Counterion density, MD calculated around duplex 271
Coupling constants, averaging 111
Coupling constants, deoxy sugar 117
Coupling constants, mole fractions and phase angles 112
Coupling constants, torsion angles 108—109
Cross-relaxation rate constants, intemuclear separation and correlation time 110
Cross-relaxation rate constants, spin diffusion 109
Crossover operations, parental solutions 237
Crosspeaks, DQF — COSY spectrum 126
Crosspeaks, intensity difference 137
Crystal conformation, pseudorotational, angle plots 157
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте


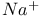 counterions, influence on gas-phase ionization potentials
counterions, influence on gas-phase ionization potentials 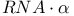 -winged ODN hybrids, denaturing gel electrophoretic analysis of RNase H digestion products
-winged ODN hybrids, denaturing gel electrophoretic analysis of RNase H digestion products  -anomeric nucleotides, base stacking and specific base pairing
-anomeric nucleotides, base stacking and specific base pairing  P NMR spectra
P NMR spectra  stretch, schematic diagram
stretch, schematic diagram  spectra of
spectra of  imino proton and methyl group regions
imino proton and methyl group regions  , backbone conformation of duplexes
, backbone conformation of duplexes