|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Leontis N.B. (ed.), SantaLucia J., Jr. (ed.) — Molecular Modeling Of Nucleic Acids |
|
|
 |
| Предметный указатель |
Crystal packing, local DNA structure 2—3
Crystal packing, structure in nucleic acid crystals 286
Crystal structure, agreement of X-ray and simulation 265
Crystal structure, B — DNA 263—265
Crystal structure, RNA 12
Crystalline DNA, particle mesh Ewald, method 286
Crystallized complexes, TBP — DNA 329
Crystallographic B factors, distribution over RNA dodecamer 75
Crystallographic studies, RNA internal, loops 56—76
Curved A-tract DNA, modeling 151—159
CURVES, calculations for helices surrounding internal loops 68
CURVES, conformational analysis for DNA 331
CURVES, Cylinder representations, comparing 3
CURVES, helicoidical parameters with respect to global helix axis 261
CURVES, irregular DNA structures 324
CURVES, parameter changes 327
CURVES, structural analysis 318
D structure, signal recognition particle RNA 405—413
Damaged DNA strand, backbone dihedral angles 321
Damaged oligonucleotides, MD simulations 312
DASGroup, Inc., The 312
Decoding site, 16S rRNA 373—374
Decoding site, key interactions with mRNA-tRNA complex 374 1
Decoding site, three-dimensional model 375
Deconvolution method, structural refinement 168
Density, high charge on array surface 223
Deoxyribose ring conformation, intraresidue spin-spin coupling patterns 97—100
Dials and Windows, conformational analysis for DNA 331
Dials and Windows, monitor conformational transitions 261
Dials and Windows, standard conformational descriptors of DNA 307
Dials and Windows, structural analysis 318
Dimethyl phosphate, conformational energies for fragments 46
Dimethyl phosphate, gauche-gauche conformer 45
Dimethyl phosphate, interaction energy with single water molecule 45
Dimethyl phosphate, selected geometrical parameters 48
Dimethyl phosphorothioate, conformational energies for fragments 46
Dimethyl phosphorothioate, interaction energy with single water molecule 45
Dimethyl phosphorothioate, model fragments 46
Dimethyl phosphorothioate, selected geometrical parameters 48 1 9
Dimethylguanine, photoelectron spectra, molecular orbital diagrams, and IPs 20
Discrimination ratios, definition 217
Distance constraints, time-averaged 112
Distance geometry, model of a pseudoknot 387
Distance geometry, nab 385—386
Distance metric, difference between two homogeneous transformation matrices 397
Distance restraints, average structure calculation 192
Distance restraints, MD refinement 144
Distance restraints, NOE-derived 131—140
Distance restraints, partial relaxation in NOESY spectra 137
Distance restraints, refinement of A-type and B-type conformers 191—193
Distance restraints, semiquantitative 124
Distance restraints, structure refinement 139
Distortion, helical axis 65—68
Distortion, intra-base-pair parameters and base-pairing properties 319
DNA antisense oligonucleotides, phosphoramidate-modified 289—291
DNA bending phenomena, accuracy of molecular dynamics 274
DNA conformational transitions, MD studies 278—280
DNA containing inverted anomeric centers and polarity reversals, structure and stability 92—105
DNA containing ring-saturated thymine, derivatives, MD simulation 318—319
DNA crystal structure, MD calculated and observed minor groove width 268
DNA crystallographic unit cell, superimposed structures from crystallography and MD simulation 267
DNA curvature, MD studies 270—278
DNA damage, molecular dynamics 291 293
DNA decamer model system,  -anomeric nucleotides 103 -anomeric nucleotides 103
DNA distortions by cis, syn thymine dimer, MD simulation 319—324
DNA duplexes, sequence-dependent structural properties 285—303
DNA fragment, irregular 324
DNA helix, TATA box-binding protein 329—345
DNA lesions, configuration 315
DNA modifications, pyrimidine bases 312
DNA mutations, mismatches 106
DNA oligonucleotide with A-tracts groove width as function of sequence 276
DNA oligonucleotide(s), pyrimidine base 312—328
DNA sequences, Raman spectra 153
DNA strand, conformation 115
DNA structural perturbations, pyrimidine lesions 313
DNA structure, localized perturbations 319 324
DNA three-way junction D NOESY-NOESY hybrid-hybrid matrix refinement 167—180
DNA three-way junction, final structure 179
DNA three-way junction, global structure after refinement 176
DNA three-way junction, refinement summary 177
DNA three-way junction, sequence 173
DNA three-way junction, structural refinement 170 174
DNA, A to B transition 278
DNA, A- and B-form geometries 42
DNA, bendability 330
DNA, canonical A and B forms 264
DNA, conditions expected to stabilize A-DNA 296—299
DNA, kinked 150 196
DNA, modeling via molecular dynamics simulation 260—284
DNA, modifications 87 88
DNA, molecular modeling 1
DNA, molecular modeling studies 150—166
DNA, pairings with modified bases 86
DNA, rms differences with respect to canonical A and B forms 306
DNA, simulations 286—288
DNA, simulations of B to A transition 306—310
DNA, spontaneous transitions between A and B forms 305
DNA-protein complex, crystallographic studies 3
DNA:RNA duplexes, structure 49—52
DNA:RNA hybrids, helical parameters 113
DNA:RNA hybrids, selected distances 115
DNA:RNA hybrids, stability and structure 288
DNAminiCarlo, generation of various conformations 185
DNAminiCarlo, potential conformers for DNA dinucleotide 186
DNAminiCarlo, rMC simulated annealing protocol 189
DNAminiCarlo, structural refinements 184
Donati, Alessandro 181
Double helix, deformation 270
Double helix, molecular modeling 1
Double helix, structural changes 3
DQF-COSY crosspeaks, control and aT duplexes and simulations 101
DQF-COSY spectrum, pyrimidine identification 126
Duplex DNA and RNA, A vs. B equilibrium in MD simulations 304—311
Duplex DNA oligonucleotides, modified pyrimidine bases 312—328
Duplex DNA, helical twist 387—391
Duplex formation, thermodynamics 206—228
Duplex oligomers, conformation 150
Duplex oligonucleotides, B/Z junctions 161—163
Duplex oligonucleotides, hydrogen bonding patterns in base pairs 77—90
Dyad symmetry, DNA diffraction pattern 1
efn2, coaxial stacking of helixes and free energy of multibranch loops 250—252
efn2, enhanced stability for coaxial stacking of adjacent helixes 252
efn2, Jacobson — Stockmayer function 251—252
Electronegative pockets, possible locations in B — DNA minor groove 273
Electrostatic interactions, importance 331—333
Electrostatic interactions, stable duplex oligonucleotide simulations 304
Empirical approaches, modeling nucleic, acid structure and dynamics 8—9
Empirical model, approximating free energy, parameters of structural motifs 247
Energy barriers, metastable and stable structure 229
Energy barriers, refolding 236—237
Energy ionization, removal of electron from phosphate 38
Energy minimization, empirical force fields 9
Energy minimized structure, stereo view 158
Ensemble approach, suitability 118
Ensemble NOEs, failure of model 119
Ensemble, structures present in solution 111—112
Environment, representation of effect on DNA 296—299
Equilibrium behavior, model 224
Equilibrium binding constants, TBP-bound, DNA 343
Equilibrium, A and B forms of DNA 296 304—311
ERNA-3D, 3D structures of signal, recognition particle RNA 405—413
Esteban, J. A. 360
Euclidian distance metric, difference between two homogeneous transformation matrices 397
European Synchrotron Radiation Facility 77
Evertsz, E. M. 150
Evolution, genetic 233
Ewald sums, lattice energy of ionic crystals 262
Ewald sums, MD simulations of nucleic acids 304
Exchangeable protons, assignments 129 131
| Exterior loops, dangling ends and coaxial stacking 253
Farr — Jones, Shauna 181
FELIX, 3D data set to give data matrix 174
Ferguson, David M. 41
Fernando, Harshica 18
Fiber diffraction, DNA molecules 1
Flexible molecules, structure determination 181
Fluorescence energy transfer, tRNA-mRNA complex 373
Fluorescence, adsorbed density 226
Fluorescence, intensity at probe sites 213
Focal elements, structural hypotheses 401
Folding algorithm, accuracy 253—255
Folding algorithm, base pairs 253
Folding algorithm, software 248
Folding dynamics, RNA 229—245
Folding pathways, relevance of GA simulations 241
Folding pathways, RNA 242 243
Folding pathways, simulated for SV 11
Folding pathways, simulations 232—233
Folding, kinetics of secondary structure formation 230—232
Force field results, AMBER 266—270
Force field results, GROMOS 265—266
Force field(s), B — RNA 288
Force field(s), empirical 8—9
Force field(s), simulations of oligonucleotides in solution 310
Force field(s), structure determination 118
Force field(s), structure of nucleic acids in solution 299—300
Forman, Jonathan E. 206
FORTRAN, RNA secondary structure, prediction 246
Free energy distribution, P:T complexes in mismatch sites 224
Free energy distribution, probe-target binding and probe-probe associations 225
Free energy landscape, GA mutations 236
Free energy minimization, secondary structure prediction 247
Free energy model, hairpin loops 249
Free energy of ionization, solution 36
Free energy penalty, closing multibranch loop 251
Free energy, phylogenetically determined base pairs 246—257
Ftouhi, Abdelmjid 394
Functional structure formation, RNA 229
Furanose ring, bent DNA 151
Furanose ring, conformation 164
Furanose ring, pseudorotational conformational wheel 154—155
Furrer, Patrick 181
Gas-phase appearance potentials, nucleotide bases 18
Gas-phase ionization potentials, influence of 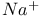 counterions 30—34 counterions 30—34
Gas-phase ionization potentials, nucleotides 24—30
Gas-phase ionization potentials, sensitivity to geometry 30
Gaussian 90
GAUSSIAN, calculations 314
Gene expression, antisense DNA therapy 92
General sequence DNA, properties 341
Genetic algorithm, computer simulations 229—245
Genetic algorithm, implementation for GA folding 235
Genetic algorithm, parameters 238
Genetic algorithm, principles 233—234
Genetic algorithm, RNA folding 234
Genetic algorithm, RNA structure prediction 247
Genetic analysis, oligonucleotide probes immobilized on solid supports 206
Germann, Markus W. 92
Ghomi, M. 150
Gibbs free energy of hydration, Langevin dipole relaxation method 34—37
Gibbs free energy of ionization in solution, evaluation 34—37
Global conformational analysis, DNA bound by TBP 339
Global curvature cis, syn thymine dimer 324
Global curvature with and without cis, syn dimer 325
Glycosidic linkage, influence on A to B equilibrium 307
Glycosidic torsion angle, variable in polynucleotides 4
Gorenstein, David G. 167
Gowda, Krishne 405
Gozansky, Elliott 167
Griffey, Richard H. 41
GROMOS, energy functions 262
GROMOS, force field results 265—266
Group I introns folding and structure of complex RNAs 355
Group I introns, helix formation 355
Gultyaev, A. P. 229
Hairpin loop, binding to SPXX-containing peptide 195—204
Hairpin loop, folding and structure formation 231
Hairpin loop, free energy model 249
Hairpin loop, interaction 355
Hairpin loop, NMR 3D structures 11
Hairpin loop, NMR spectra of oligonucleotide d(T6C4A6) 197—199
Hairpin loop, nucleation points for folding 355
Hairpin loop, RNA structure 293
Hairpin loop, RNA — RNA anchors 355
Hairpin loop, sequence and numbering of residues 198
Hairpin loop, stability of closing base pair and first mismatch 250
Hairpin motif, NMR study 123—124
Hairpin ribozyme and cognate substrate, secondary structure of complex 362
Hairpin ribozyme, kinetic mechanism 365
Hairpin ribozyme, reengineering for homogeneous folding 366—367
Hairpin ribozyme, structure and dynamics 360—368
Hammerhead ribozyme, ribbon drawing 349
Hammerhead ribozyme, three-way junction with two single-stranded stretches 352
Harvey, Stephen C. 369
Heckman, J. E. 360
Helical axes, distortion 70
Helical conformation, determination 108
Helical distortion, internal loops 65—68
Helical sections, distances 410
Helical twist, B — DNA structures 3
Helical twist, duplex DNA 387—391
Helical twist, sequence dependence 288
Helices, asymmetry 352
Helices, coaxial stacking 252
Helicoidal parameters, backbone/sugar torsions 381
Helicoidal parameters, variation 116
Helix 4, stabilization 363
Helix zipping, interactions for short oligonucleotides 206
Hepatitis delta virus, pseudoknotted three-way junction 351
Heteronuclear NMR methods, labeled RNA 124
Holbrook, Stephen R. 56
Homogeneous transformation matrices, conformational search space size 397
Homogeneous transformation matrices, contact between nucleotides 396
Homogeneous transformation matrices, spatial information 396
Homonuclear 2D NOE spectroscopy, non-exchangeable proton resonances 144
Homonuclear 3D NOESY-NOESY, spectroscopy, structure of large biomolecules 167
Hoogsteen base pairing, DNA and RNA 2
Hunter, William N. 77
Hybrid-hybrid relaxation matrix refinement 3D NOESY — NOESY data 167—180
Hybrid-hybrid relaxation matrix refinement DNA three-way junction 170 174
Hybrid-hybrid relaxation matrix refinement structural refinement 169
Hybrid-hybrid relaxation matrix refinement, advantages 176
Hybrid-hybrid relaxation matrix refinement, method accuracy 180
Hydration, A — DNA stabilization 299
Hydration, energetic ordering of ionization events 35—37
Hydration, ionization energies of nucleotides 35 38
Hydration, local variation in B-DNA crystal structures 3
Hydrogen bond, constraints 174
Hydrogen bond, definition 79
Hydrogen bond, distance, lesion base pairs 292
Hydrogen bond, mRNA-tRNA complex 376
Hydrogen bond, thermodynamic stability of UNCG class of RNA tetraloops 295
Hydrogen bonding patterns, base pairs of duplex oligonucleotides 77—90
Hydrogen bonding restraints, molecular dynamics 140
Hydrogen-bonded DNA bases, high-resolution structure 2
Illangasekare, Nishantha 167
Imino 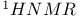 , base pairing in nucleic acids 97 , base pairing in nucleic acids 97
Independently folding domains, ribozyme structures 363
Inhibition of replication, pyrimidine DNA lesions 313
Insight II, overlay procedure 331
Insight II, structures of DNA:RNA hybrid 113
Intensity differences, limitation for weak peaks 137
Interfacial environment, DNA duplex formation 221—223
Interhelical angle and displacement in helices, calculation 69
Intermolecular distances, probe array 208
Intermolecular RNA-RNA complexes, role of internal loops in stabilization 59
Internal bulges, variable flexibility 355
Internal curvature, curved helical axis 65
Internal loops, distortion and sequence 56
|
|
 |
| Реклама |
 |
|
|
 |
 |
|
 |
|
О проекте
|
|
О проекте


 -anomeric nucleotides
-anomeric nucleotides 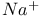 counterions
counterions 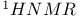 , base pairing in nucleic acids
, base pairing in nucleic acids