|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Leontis N.B. (ed.), SantaLucia J., Jr. (ed.) — Molecular Modeling Of Nucleic Acids |
|
|
 |
| Предметный указатель |
Synthesis sites, melting curves 225
Synthesis sites, probe distributions 222
Tan, Robert K. — Z. 369
Tandem mismatches, free energy parameters 248
Target binding, length dependence 217
Target duplex, Cartesian RMSD of refined structures 171
Target molecules, bridge between two probes 223
Target molecules, structures 114
Target oligonucleotides, sequences 221
TATA box, protein partner selection 329—345
TATA box-binding protein binding sites 330
TATA box-binding protein, DNA helix 329—345
TATA box-binding protein, RNA polymerase 329
Tautomeric form, nucleotide bases 1—2
TBP-bound DNA, local conformational properties and analysis 339—341
TBP-bound DNA, transient properties 341—342
Tectonics, RNA 346—358
Tetraloops, enhanced stability 249
Tetraloops, GnRA family 126
Tetraloops, RNA 293—295
Tetraloops, stability bonuses 251
Thermally bound state, macromolecular MD simulation 261
Thermodynamic parameters RNA folding 248
Thermodynamic parameters RNA secondary structure prediction 246—257
Thermodynamic properties,  -containing DNA duplexes 94 -containing DNA duplexes 94
Thermodynamic stability RNA helices 352
Thermodynamic stability sequence dependent 255
Thermodynamic studies, RNA folding and DNA metabolism 9—10
Thermodynamic(s), duplex formation and mismatch discrimination 206—228
Thiviyanthan, Varatharasa 167
Thomas Jefferson University 92
Three-dimensional structure, large RNA molecules 6
Three-way junctions, single-stranded region 354
Thymidine glycol, stereoisomers 315
Thymine base damage, distortions of oligonucleotide 318
Thymine carbonyl vibrations in D 20
Thymine glycol in DNA, cis isomers 317
Thymine lesions, ring-saturated 313—318
Thymine, configuration 315
Thymine-derivative stereoisomers, relative stability 314
Tobacco mosaic virus, pseudoknotted three-way junction 351
Torsion angles, nucleic acid backbones 51—52
Torsion angles, parameters 107
Torsion angles, semiquantitative distance restraints 124
Torsion angles, variation 116
Torsional energy profile, nonbonded parameters 44 45
TpA steps, flexibility in response to changes 319 324
Transcription rate, accurate formation of functional RNA structures 231
Transformation(s), nab 384
Transformational set, homogeneous transformation matrices 396
Transition mechanism, B-DNA to A-DNA 297—299
Transition mutations, guanine reaction with alkylnitrosoureas 86
Translation, antibiotic bound to decoding site 376
Triangle smoothing, distance geometry 386
Triloops, enhanced stability 249
Triloops, stability bonuses 251
tRNA, adapter molecules 5
tRNA, clover-leaf model 5 6
tRNA, ribbon diagram 353
Trulson, Mark O. 206
Truncated probes, broadening and depression of melting curves 224
Truncated relaxation effects, correction 135
Turner, Douglas H. 246
Two-spin approximation, structural refinement 167—168
Two-spin approximation, systematic errors 168
Ultraviolet radiation, photocrosslink 374
Ulyanov, Nikolai B. 181
Uncertainty principle, terminology and notation 398—399
United atom representation, macromolecular systems 370
| Universidad Autonoma de Barcelona 329
Universite de Montreal 394
Universite Pierre et Marie Curie 150
University of Alabama at Birmingham 369
University of Calgary 92 195
University of California at Berkeley 56
University of California at San Francisco 122 181 285
University of Dundee 77
University of Illinois at Chicago, The 18
University of Maryland at Baltimore 304
University of Minnesota 41
University of Oregon 150
University of Rochester 246
University of San Francisco 285
University of Southampton 77
University of Texas Health Science Center 405
University of Texas Medical Branch 167
University of Vermont 360
University of Warsaw 285
Uracil, lowest energy k and lone-pair ionization potentials 22
UV-induced mutations, dipyrimidine sequences 313
Valence electronic structure, nucleotides 18
van Batenburg, F. H. D. 229
van de Sande, Johan H. 92
van der Waals parameters 43—44
VanLoock, Margaret S. 369
Variable-length linkers, coaxial stacking of helices 363
Viroid RNA, conserved metastable hairpin 244
Viroid RNA, metastable hairpins for efficient replication 231
Viroid(s), photoreactive motif within internal loop B 361
Vogel, Hans J. 195
Walter, N. 360
Walton, Ian D. 206
Washington University 246
Water activity, DNA conformational transitions 278
Water activity, stabilization of A — DNA 296—297
Water docking, dimethyl phosphate and dimethyl phosphorothioate models 44
Water structure, base pairs in internal loops 71
Water-counterion environments, energetics of nucleotide ionization 18—40
Watson-Crick base pairs,  and and  78 78
Watson-Crick base pairs, complementarity 77—79
Watson-Crick base pairs, duplex structures 77
Watson-Crick base pairs, hydrogen bond stabilization 79
Watson-Crick base pairs, identification 129
Watson-Crick base pairs, isosteric in antiparallel helices 347
Watson-Crick base pairs, sequence and conformational junctions 161
Watson-Crick hydrogen bonds base pairs near cis-syn thymine dimer 323
Wayne State University 1
Weinstein, Harel 329
Wesleyan University 260
Westhof, Eric 346
Wobble configuration, purine-pyrimidine, pair 80
Wobble pair, NH protons 129
Wormlike chain model, DNA 12
X-PLOR, molecular modeling problem 380
X-PLOR, pseudoknot 386
X-PLOR, starting model coordinates 174
X-ray crystal structures, RNA internal loops 56—76
X-ray fiber diffraction analysis, RNA samples 6
X-ray fiber diffraction data, B-form DNA 2
yammp, molecular modeling problem 380
Yeast  , ribbon diagram 353 , ribbon diagram 353
Yeast  , , 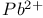 cleavage of specific ribophosphodiester bond 402 cleavage of specific ribophosphodiester bond 402
Young, M. A. 260
Z DNA, alternating cytosine and guanine 159
Zhou, Ning 195
Zhu, Frank 167
Zuker, Michael 246
Zwieb, Christian 405
Zwitterion radicals, base ionization 37
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте


 -containing DNA duplexes
-containing DNA duplexes  and
and 
 , ribbon diagram
, ribbon diagram  ,
, 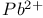 cleavage of specific ribophosphodiester bond
cleavage of specific ribophosphodiester bond