|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Leontis N.B. (ed.), SantaLucia J., Jr. (ed.) — Molecular Modeling Of Nucleic Acids |
|
|
 |
| Предметный указатель |
Internal loops, free energy parameters 248
Internal loops, helical distortion 65—68
Internal loops, hinge effect 141 43
Internal loops, nucleotide RNA 123
Internal loops, RNA secondary structure 56—76
Internal loops, structural stabilization 129
Internal loops, X-ray crystal structures 59—65
Interproton distances, calculation from experimental NOE intensities 184
Interproton distances, NOE intensities 187
Intra-base-pair parameters, simulated with cis, syn dimer 322
Intramolecular cleavage, prevention 363
Intron P4—P6 domain, internal loops in crystal structure 65 67
Inverted anomeric centers, DNA structure and stability 92—105
Ion atmosphere, concentration dependence and interactions with DNA 270
Ion pairs, evidence for problems in handling electrostatics 333
Ionization energy, nucleotides 18 40
Ionization potentials, calculated 25 26
Ionization potentials, energetic ordering 28
Ionization potentials, nucleotide components 19—24
Ionization potentials, order 30
Ionization potentials, valence electron 27 29
Ionization, formation of zwitterion radical 37
Ionizing radiation, DNA damage 37
ISIS Pharmaceuticals 41
Iterative refinement process, structure 174—176
J-coupling data, nucleotides 100
Jacobson-Stockmayer function, multibranch loops 251—252
Jaeger, Luc 346
James, Thomas L. 122 181
JUMNA, constraint satisfaction problem 381
Junctions, formed by converging helices 352
Karplus equation, accuracy of torsion, angles 109
Kim, James 246
Kim, Nancy S. 18
Kinetic analysis, pre-steady-state 363—364
Kissing complex, interaction between two RNA molecules 355
Kollman, P. A. 285
Konerding, David E. 181
Lane, Andrew N. 106
Larsen, Niels 405
Leadzyme, application of structural hypotheses 402—403
LeBreton, Pierre R. 18
Leiden University 229
Lemieux, Sebastien 394
Leonard, Gordon A. 77
Leontis, Neocles B. 1 167
Lesion sites, photodamaged decamers 291—293
Lesion sites, torsion angles 292
Lesion(s), ring-saturated and thymine-dimer 326
Ligand binding, RNA internal loops 68—74
Ligation reactions, kinetics 364
Limb-joint approach, RNA secondary structures 407
Lind, Kenneth E. 41
Linked-atom methods, polynucleotide 2
Liu, He 181
Local axis deformation, MD model 265
Local referential, nucleotide 396
Long-range interactions, computation of forces in MD 262
Loop energies, preference for short-range weak stems 236
Loop hydroxyls, atomic-level interactions 295
Loop size, destabilizing loop contribution 230
Loop-loop motif, pseudoknot 355
Looped-out bases, bulging of individual bases 74
Lukavsky, Peter 122
Luxon, Bruce A. 167
Macke, Thomas J. 379
MacKerell, Alexander D., Jr 304
Macromolecular models, level of resolution 369
Magnetization transfer, efficiency 10
Major Francois 394
Major groove, A — DNA stabilization 299
Mapping, active variants and structural hypotheses 399
MARDIGRAS, accurate distance bounds from NOE data 186
MARDIGRAS, complete relaxation matrix calculations 131
MARDIGRAS, dipole-dipole relaxation rates 184 185
MARDIGRAS, distance restraints 139—141 189
MARDIGRAS, interproton distances 124—125
MARDIGRAS, isotropic motional model 139
MARDIGRAS, partial relaxation correction 137
Marker band, B-form DNA 154
Masquida, Benoit 346
Match signal, ratio to mismatch signal 219
Mathews, David H. 246
Max Planck Institute for Molecular Genetics 405
MC-SYM, model of decoding site 373
MC-SYM, molecular modeling problem 380
MC-SYM, transform structural data into 3D models 395—404
MD calculated average structures 275
MEDUSA, repeated refinements against internally consistent subsets of restraints 182
MEDUSA, results of conformer ensemble refinement 189—193
Melting behavior, base composition and sequence 224
Melting curves, matched and single-base mismatched probes 218
Melting curves, matched probe synthesis sites 216
Melting curves, matched probe(s) 220
Melting curves, oligonucleotide adsorption 206
Melting curves, role of probe length heterogeneity 221
Melting curves, simulated vs. experimental 221
Melting curves, structure 213
Melting curves, target concentrations 213—217
Metal ions, crystal structures of RNA oligomers 72
Metal-assisted RNA catalysis, classical model 402
Metastable structures, revealed by GA 241—244
Methyl uracils, lowest energy 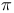 and lone-pair ionization potentials 22 and lone-pair ionization potentials 22
Metric matrix distance geometry, nab 385—386
Metropolis Monte Carlo, structural, refinements 184
Mfold, folding algorithm 248
Miaskiewicz, Karol 312
Michigan State University 405
Miller, J. L. 285
Miller, John 312
Minor groove binding, sequence specificity 195—196
Minor groove, A — DNA stabilization 299
Minor groove, A-tract DNA 159
Minor groove, ApT pocket 270
Minor groove, Biograf 156
Minor groove, DNA:RNA duplexes 49
Minor groove, extended conformation 204
Minor groove, hybrid duplex 42
Minor groove, local variation in B-DNA crystal structures 3
Minor groove, TBP binding site 329—330
Misfolding, wild type ribozyme 363
Mismatch(es) base pairs 106
Mismatch(es) coaxial stacking 252
Mismatch(es) crystallographic study 80
Mismatch(es) internal loop 248
Mismatch(es) melting temperature depression 206
Mismatch(es) Mismatch discrimination, thermodynamics 206—228
Mismatch(es) on temperature, concentration, and probe length 217
Mismatch(es) purine-purine base pairs 80—82
Mismatch(es) pyrimidine-pyrimidine base pairs 82—86
Mismatch(es) Watson — Crick base pairs 79—80
Mismatched probe sites, adsorption rates for densities of adsorbed target 213
Mispair formation, error occurring on replication 80
Mixed-resolution models, motivation 371 372
Mlp 2D root mean square distance plot 339
Mlp production phase simulation 331
Mobility parameters, spherical distribution of displacement 74
Modeling, 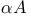 and control decamer duplexes 100—103 and control decamer duplexes 100—103
Modeling, curved A-tract DNA 151—159
Modeling, RNA 4—7
Modified DNA:RNA hybrid, RNase H activity 42—43
Modular modeling of RNA, RNA tectonics 346—358
Modular units, modeling of macro-molecules 350
Modularity, RNA structure 357
Mohan, Venkatraman 41
Molecular computation languages, conformational searches 380
Molecular contacts, conformational search space 396—397
Molecular dynamics simulation, description 260—263
Molecular dynamics simulation, DNA modeling 260—284
Molecular dynamics simulation, duplex DNA or RNA in solution 304—311
| Molecular dynamics simulation, normal dodecamer and NP-modified sequence 290—291
Molecular dynamics simulation, nucleic acid systems 285—303
Molecular Dynamics Tool Chest, analysis of MD on DNA and proteins 261
Molecular dynamics, results of simulations 50
Molecular dynamics, time-average and restrained 182
Molecular dynamics, trajectory analysis 261
Molecular force field, analytical energy function 262
Molecular loops, nab 382
Molecular mechanics, nab 386
Molecular modeling language, OCL 381
Molecular modeling of DNA, Raman and NMR data 150—166
Molecular modeling, tools 12—13
Molecular motion, biological functioning 11
Molecular orbital diagrams, calculated 26
Molecular simulations, AMBER4.1 package 48—49
Monte Carlo procedure, favorable stem combinations 232—233
Monte Carlo procedure, low-energy dinucleotide conformers 380
MORASS, restrained MD iterative refinement cycles 174
MORCAD, object transformations 381
Mosaic modeling, RNA 347 350
Motif swap, RNA motifs 357
Mount Sinai School of Medicine 312 329
mRNA, protein synthesis in cells 5
Mujeeb, Anwer 181
Muller, Florian 405
Multibranch loops, free energy 250—252
Multiple conformations, nucleotides 111—112
Multiple molecular dynamics method, sampling in RNA simulations 295
Mutagenesis, pyrimidine DNA lesions 313
Mutagenic pathways, Watson-Crick base pairs 79—80
Mutation, base pairing 86
Mutation, compensatory base changes 406
Mutation, iteration of the GA 237
Mutational operations, selection of fittest solutions 233
Nab, construction of 3D molecular models 379—393
Nab, design 382—384
Nab, examples of use 386
Nab, potential applications 391
National Institute for Medical Research 106
National Institutes of Health 285
Natural systems, rate of RNA folding 231 232
Nearest neighbor free energy parameters, predicting secondary structure 247
NEW HELIX '93, propeller twisting values 156 159
Newhelix, local axis convention for base pair steps 261
NMR data, determination of structural ensembles 181—194
NMR spectroscopy, solution structure 10—11
NMR structure determination, tools 183 184
NMR, nucleic acid structures in solution 106—121
NMR, sample preparation and experiments 145—146
NMR, studies of a-containing DNA duplexes 97
NOE build-up rate equation, structural refinement 168—169
NOE integrals, per residue 178
NOE intensities, calculation from structures 183—184
NOE intensities, structure refinement calculations 137 139
NOE time courses, glycosidic torsion angles 112
NOE-derived distances, multispin effects 124
NOESY walks, nonlabile  resonances in B — DNA 97 resonances in B — DNA 97
NOESY — NOESY volumes, simulated 171
Non-Watson-Crick base pairs, internal, loops 59
Nonexchangeable protons, assignments 125—129
Nuclear Overhauser effect, adduct 292—293
Nuclear Overhauser effect, buildup curves 265—266
Nuclear Overhauser effect, interproton distance for 6—4
Nuclear Overhauser effect, relaxation matrix methods 122—149
Nuclear RNA polymerases, TATA box-binding protein for transcription 329
Nucleic acid complexes, physical properties 41—54
Nucleic Acid Database, RNA and DNA structures 7
Nucleic acid structures, modeling 379—393
Nucleic acid structures, reasonable representation 286—288
Nucleic acid(s), conformational analysis 106—121
Nucleic acid(s), probe immobilized on surface 207
Nucleic acid(s), pseudoatom representation 370—371
Nucleic acid(s), simulation instability 286—287
Nucleic acid(s), structural parameters 106—108
Nucleic Acids Data Bank, experimental data for crystal structures 263
Nucleosides, distribution in rRNA internal loops 60
Nucleosome core particle, schematic 389
Nucleosome model, nab 387—391
Nucleosome model, program 390
Nucleotide bases, tautomeric form 1—2
Nucleotide contact graph, spanning tree 395
Nucleotide counterion clusters, ionization energies 38
Nucleotide Gibbs free energies of activation, aqueous bulk solvation 34—37
Nucleotide ionization potentials, clusters containing Na+ bound to phosphate and H 20 33
Nucleotide ionization, energetics in water-counterion environments 18—40
Nucleotide(s) gas-phase ionization potentials 24—30
Nucleotide(s) lowest free energy structure 255
Nucleotide(s) structural parameters 107—108
Occupied orbitals, calculated 25
OCL, objects’ positions and orientations 381
Oil drop model, DNA conformational transitions 278
Oligomers, Raman spectra 152
Oligonucleotide arrays, photolithographically synthesized 206—228
Oligonucleotide interactions, in solution and at interface with surface-bound probes 209
Oligonucleotide simulations, review 305
Oligonucleotide(s) design of antisense therapeutics 92—105
Oligonucleotide(s) DNA configuration 2
Oligonucleotide(s) hydrogen bonding patterns in base pairs 77—90
Oligonucleotide(s) pyrimidine base 312—328
Oligonucleotide(s) sequences 208
OPLS, force fields with balanced solute-solvent and solvent-solvent interactions 286
Optimization calculations, partial geometry 31 3
Osman, Roman 312
Pacific Northwest Laboratory 312
Papadantonakis, George A. 18
Parameterization, phosphorothioate-DNA:RNA hybrids 43—48t
Pardo, Leonardo 329
PARSE, pool of potential conformers 182—183
PARSE, results of conformer ensemble refinement 189—193
Partial relaxation 2D NOE spectra 138
Partial relaxation effects, correction 135 137
Particle mesh Ewald algorithm, electrostatic interactions 331
Particle mesh Ewald algorithm, lattice energy of ionic crystals 262—263
Particle mesh Ewald electrostatics, charged systems like nucleic acids 286
Particle mesh Ewald electrostatics, electrostatic potential energy 319
Particle mesh Ewald electrostatics, MD simulations on nucleic acid systems 285—303
Particle mesh Ewald method, conformational behavior of oligo-nucleotides in aqueous solution 289
Paster, Nina 329
PDQPRO, correct structures and probabilities 186
PDQPRO, equilibrium of two conformations 193
PDQPRO, global minimum of quadratic function 185
PDQPRO, reconstitute synthetic structural ensembles 185—186
Peptide from HMW basic nuclear proteins, binding to A-T rich DNA hairpin loop 195—204
Peptide sample, NMR titration of aromatic, amide, and imino protein regions 200 201
Peptide, interaction with hairpin C loop region 204
Peptide-DNA sample, characterization of changes 199
Periodic box simulations, PME model 286
Periodicity, effect on neighboring cells 287
Persistence length index, statistical theory of chain molecules 261
Perturbation energies, combining nucleotide components 23—24 1 10
Peticolas, W. L. 150
Pharmacologiques et Toxicologiques 150
Phenanthroline-copper ion, nuclease activity 159—166
Phosphates, backbone structure 42
Phosphates, new parameters 47
Phosphodiester backbone, A vs. B equilibrium 310
Phosphoramidate analogs, conformational properties 289
Phosphoramidate analogs, stable duplexes and triplexes 289
Phosphoramidate-modified DNA antisense oligonucleotides 289—291
Phosphorothioate backbone, energy surface 52
Phosphorothioate nucleic acids, parameterization and simulation of physical properties 41—54
Phosphorothioate oligonucleotides, structural characteristics 52
Phosphorothioate(s) antisense therapies 42—43
Phosphorothioate(s) backbone structure 42
Phosphorothioate(s) new parameters 47
Phosphorothioate-DNA:RNA hybrids, melting temperatures 52
Phosphorothioate-DNA:RNA hybrids, parameterization 43—48
Phosphorothioate-DNA:RNA hybrids, structure and activity 42—43
Photocrosslink, UV radiation 374
Photocrosslinking efficiency, RNA loops 361
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте


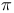 and lone-pair ionization potentials
and lone-pair ionization potentials 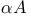 and control decamer duplexes
and control decamer duplexes  resonances in B — DNA
resonances in B — DNA