|
|
 |
| Авторизация |
|
|
 |
| Поиск по указателям |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Leontis N.B. (ed.), SantaLucia J., Jr. (ed.) — Molecular Modeling Of Nucleic Acids |
|
|
 |
| Предметный указатель |
Photodamaged DNA in aqueous solution, molecular dynamics 291—293
Photoelectron spectroscopy, valence manifold IPs of intact nucleotides 23
Photolithographic synthesis, oligonucleotide arrays 206—208
Photolithographic synthesis, probe array preparation 206
Phylogenetic analysis, structural motifs 12
Phylogenetic methods, model 3D structures 6
Phylogenetic structure, free energy minimization 255
Phylogenetic structure, scoring against base pairs 253
Phylogenetically determined base pairs, free energy 246—257
Plasmid replication, metastable folding 244
Plasticity, oligonucleotides 106
Pleij, C. W. A. 229
Pockets, localized complexation of otherwise mobile counterions 270
Polar groups in equatorial plane, interactions 315
Polarity reversals, DNA structure and stability 92—105
Poly(dA)*(poly(dT), phosphate-deoxy-ribose ring vibration 151
Polynucleotides, conformational analysis 4
Potential conformers, probability assessment 185—189
Potential conformers, probability distribution 182—183
Predictions, final RNA structure 238
Probability assessment, factors 186
Probability assessment, folding steps 234—236
Probability assessment, structural ensembles 181—194
Probe array, density 223
Probe array, difference from bulk aqueous reference system 217
Probe array, electrostatic factors 223
Probe array, features 210
Probe array, labeled target oligonucleotides 207
Probe bridging, target DNA binding 206
Probe length, dependence on adsorption densities 213 217
Probe length, free energies for P:T binding 224
Probe length, rates of adsorption 211—213
Probe(s) interactions 223
Probe(s) recognition of DNA or RNA from solution 206
Probe-probe interactions, stability of probe-target duplexes relative to solution 208
Propeller twist, base pairs 158
Propeller twist, bent DNA 151
Propeller twist, Biograf 156
Propeller twist, MD model 265
Protection patterns, drugs 373
Protein coding genes, TATA box 329
Protein partner, TATA box selection 329—345
Protein recognition and binding, internal loops 57—59
Protein synthesis, sensitivity to time of folding 231—232
Protein translocation, endoplasmic reticulum membrane 123
Protein-DNA interactions, sequence discrimination 195
Proton assignment, strategy 125—129
Proton chemical shift, assignments 128
Proton-proton dipolar interactions, DNA 107
Proton-proton scalar coupling constants, conformation of sugar rings 181
Prototype B-DNA oligonucleotide, MD studies 263—265
Pseudoatom representation, large nucleic acids 370—371
Pseudoknots, either secondary or tertiary structure 352 355
Pseudoknots, molecular mechanics 386
Pseudoknots, predicted in plant viral RNAs 239 240
Pseudoknots, predictions 238—241
Pseudoknots, program 388
Pseudoknots, RNA structure 12
Pseudoknots, single-stranded RNA 387
Pseudoknots, tRNA 350
Pseudorotational conformational wheel, furanose ring 154—155
Pseudorotational phase angles, distribution in conformers selected 190
Pucker restraints, MD refinement 144
Pucker restraints, molecular dynamics 140
Puckering, furanose ring 154
Puckering, nucleic acid studies 49—52
Puckering, phosphorothioate strand 52
Puckering, sugar ring 4 (see also Sugar puckering Sugar
Purine-purine base pairs, mismatches 84
Purine-purine base pairs, variable pairing 83 84
Purine-purine base pairs, variation and mismatches 80—82
Purine-pyrimidine base pairs, mismatch 80
Purine-pyrimidine base pairs, wobble configuration 81
Pyrimidine and purine nucleotides, sugar pucker phase angle and glycosidic bond torsional angle 336
Pyrimidine-pyrimidine base pairs,  mispair 85 mispair 85
Pyrimidine-pyrimidine base pairs, mismatches 82—86
Pyrimidine-pyrimidine base pairs, two forms of  pairing 85 pairing 85
QUANTA, initial DNA structures 331
Quantum mechanical treatments, chemical and biological properties of nucleic acids 7—8
Radial distribution functions, ionic species 332
Raman bands, A-tract DNA 151—155
Raman measurements, carbonyl vibrations 159
RANDMARDI, error analysis option 184
Rapid internal motions, nucleic acids 110—111
Rasterwood, Thomas R. 369
Rava, Richard P. 206
Recognition motifs, RNA 355
Recursive algorithm, RNA secondary structure prediction 246—257
Recursive algorithm, sequence dependence 255
Recycling delays 2D NMR 135
Reduced models, double stranded DNA 1—12
Relaxation matrix, assumed ensemble 118
Relaxation matrix, corrected NOE intensities 122—149
Relaxation matrix, derivation methods 110
Relaxation matrix, model structure 139
Repair enzyme action, determinants 293
Replication fidelity, protein recognition and repair system 79
Replication fidelity, Watson — Crick base pairs 77
Replication of RNA molecules, ribozyme 360
Research Institute of Scripps Clinic 379
Restraint generation, refinement procedures 146
Ribosomal decoding site, atomic resolution models 372—374
Ribosome, molecular modeling studies 369—378
Ribosome, uncertainty of low-resolution model 371
Ribozyme, multiple conformations 361
Ribozyme-substrate complex in vitro selection 361
Rigid body transformations, model structures 384
Ring pucker, curvature of DNA 150
Ring-saturated pyrimidine derivatives, degenerate enantiomers 314
Ring-saturated pyrimidine lesions, biological consequences 313
Ring-saturated thymine derivatives computation methods 326
Ring-saturated thymine derivatives conformational preferences 313—318
Ring-saturated thymine derivatives time sequence of the rise parameter 320
Ring-saturated uracil derivatives, interaction between pseudoequatorial substituents 316
Ring-saturated uracil derivatives, rMD calculations, statistics 117
Ring-saturated uracil derivatives, rms differences, A and B forms of DNA 308 339
Ring-saturated uracil derivatives, rmsd values, between structures 113
RMS(t), canonical A-DNA and B-DNA 279
RNA dodecamer duplex, structure 60
RNA double helices, internal loops 56
RNA folding dynamics, computer simulations by genetic algorithm 229—245
RNA folding dynamics, physical-chemical features 229—232
RNA folding, 3D architecture 347
RNA folding, implementation of principles of genetic algorithm for simulation 233—238
RNA folding, kinetic features 241
RNA folding, Monte Carlo simulations 232—233
RNA homologues, SRP species and phylogenetic comparisons 123
RNA internal loops, crystallographic studies 56—76
RNA internal loops, definition 58
RNA internal loops, flexibility 74
RNA internal loops, functions 57—59
RNA internal loops, ligand binding 68—74
RNA mosaic units, assembly rules 350—355
RNA motif, determination 123
RNA oligomer crystals, internal loops 59—65
RNA oligomer(s) non — Watson — Crick base pairs 66
RNA oligomer(s) solvent-accessible surface representations 64
RNA oligomer(s) structure stereoviews 61 63
RNA polymerase, TATA box-binding protein for transcription 329
RNA secondary structure 360
RNA secondary structure, hierarchy in formation 230
RNA secondary structure, prediction by recursive algorithm 246—257
RNA strand, conformation 115
RNA structure, optimal and suboptimal 232
RNA structure, prediction by simulation of folding pathways 232—233
RNA targets, RNase H degradation 96
RNA targets, substoichiometric destruction 94
RNA tectonics, modular modeling of RNA 346—358
RNA tetraloop, incorrect and correct loop conformation 293—294
RNA, 3D computer modeling from low-resolution data and multiple-sequence information 394 404
| RNA, A-form geometry 42
RNA, barriers to structural interconversion 294
RNA, kinetics of secondary structure formation 230—232
RNA, mosaic modeling 350
RNA, mosaic structure 347
RNA, secondary structure and overall architecture 346
RNA, secondary structure determination 347
RNA, secondary structure elements 348
RNA, secondary structure models 5
RNA, simulation of folding as study tool 238—244
RNA, simulation(s) 286—288
RNA, synthesis and purification 145 (see also mRNA tRNA)
RNA-RNA anchors, hairpin loops 355
RNA-RNA anchors, schematic drawings 356
RNase H activity, antisense ODNs 103
RNase H studies, activation 92—94
Root mean square deviation, stability and convergence behavior of simulations 266
Root mean square deviations vs. time, MD structures 261
Root mean square distance, time evolution 340
rRNA, photoreactive motif within internal loop B 361
Rush, T. S., Ill 150
Salt concentration, A vs. B equilibrium 310
Salt concentration, bending behavior 152
Salt concentration, Raman band intensity 161
Sampling, multiple molecular dynamics method 295
SantaLucia, John, Jr 1
Sargueil, B. 360
Saturating adsorption densities, match vs. mismatch 224
Scalar coupling data, equilibrium of sugars 181—182
Schmitz, Uli 122 181
Schrodinger equation, approximations 7
Secondary structure, base paired helices 355
Secondary structure, RNA 347
Secondary structure, signal recognition particle RNA 406
Selectivity determinants, DNA bendability 343
Selectivity determinants, relations to measured affinities 343
Selectivity determinants, TBP-bound DNA 342
Sequence dependence recursive algorithm 255
Sequence dependence recursive algorithm selected distances in 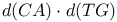 187 187
Sequence design, SRP RNA domain IV 125
Sequence junctions, nonstraight DNA 159 163
Sequence occurrence, approximating free energy parameters of structural motifs 247
Sequence specificity hydrogen bonding 196
Sequence specificity major-minor groove binding proteins 195
Sequence specificity TBP 330
Sequence(s), insight into thermodynamic and structural effects 94
Sequence-dependent variations, local structure of DNA 2
Sequence-directed curved DNA, models 151
Sequence-specific bending patterns, DNA decamer 287
Sequence-structure relation, calculating possibilities 399—402
Sequence-structure relation, patterns of base pairing and stacking 397—398
Sequential assignments, protons of a 28-mer RNA 122
Serine phosphorylation, modulation of DNA binding 196
SHAKE, constraining all hydrogen atoms 290
Sherlin, Luke D. 41
Signal recognition particle RNA, 3D comparison of small domain 411
Signal recognition particle RNA, coaxially oriented helices 407 410
Signal recognition particle RNA, common nomenclature 407
Signal recognition particle RNA, comparative modeling of 3D structure 405—413
Signal recognition particle RNA, function 405
Signal recognition particle RNA, mammalian 123
Signal recognition particle RNA, NMR structure determination of 28-nucleotide RNA 122—149
Signal recognition particle RNA, outlook 412
Signal recognition particle RNA, protein interactions and assembly of ribonucleoprotein particles 405
Signal recognition particle RNA, secondary structures 408 409
Signal recognition particle RNA, structural and functional properties of molecules 405
Signal recognition particle RNA, structure 412
Signal recognition particle RNA, tertiary interactions 407
Simulaid, simulation cells trimmed to hexagonal prisms 331
Simulation protocol, test and development 287
Single-turnover cleavage, kinetics 364
Small domain, additional nucleotides 412
Solution structure, determination 183
Solution structure, NMR spectroscopy 10—11
Solvation, ionization of nucleotides and nucleotide clusters 34—37
Solvent binding sites, crystal structures 72
Solvent binding sites, internal loop 73
Spanning tree, building procedure 397
Spanning tree, nucleotide contact graph 395
Spassky, A. 150
Spatial information, homogeneous transformation matrices 396
Spatial relation, two nitrogen bases 396
Spector, T. I. 285
Spectral overlap 2D NOESY spectra 167
Spin diffusion, biomolecules 10
Spin diffusion, estimate of cross-relaxation rate constant 109—110
Spin-spin splittings, orientation of vectors with respect to global axis 120
Spine of hydration, minor groove 270
Spine of hydration, ordered network of solvent peaks in  density 263 density 263
Sprous, D. 260
SPXX-containing peptide, binding to DNA hairpin loop 195—204
SRP 28 mer 1D NMR spectra of aromatic region 127
SRP 28 mer 2D NOE data 132f—133f
SRP 28 mer convergence of restrained MD refinement procedure 142
SRP 28 mer sequential assignments 130f
SRP 28 mer stereoviews of final rMD structure 143f
SRP 28 mer T1 relaxation times 136
SRP domain IV, sequences and phylogenetically derived secondary structures 127
Stability bonuses, tetraloops and triloops 249 251t
Stability, oligonucleotides and synthetic polymers 5
Stability, stable simulation of metastable states 300
Stability, stacked pairs 8
Stability, surface-bound duplexes 208
Stabilization, A — DNA 299
Stacking energy, base pairs at helical interface 365
Stacking energy, rate of stem melting 232
Stacking interactions, determination 141
Stem formation, energy barrier of loop 230
Stem formation, Monte Carlo procedure 234—236
Stem formation, simulation 232
Stem(s), disruption 236
Stereochemical reasoning, double helical model 1
Stern, David 206
Structural analyses, oligonucleotide starting conformations 49—52
Structural data, theory vs. experiments 333
Structural ensembles, determination from NMR data 181—194
Structural hypotheses, mathematical model based on fuzzy logic 403
Structural parameters, minimum needed for determination 106—108
Structural refinement, deconvolution method 168
Structural refinement, hybrid-hybrid matrix method 169
Structural refinement, NOE build-up rate equation 168—169
Structural refinement, optimization of conformational energy and penalty function 184
Structural refinement, restrained molecular dynamics 140—144
Structural transition, A to B form of DNA 307
Structure creation, methods 384—386
Structure determination, procedure overview 134
Structure determination, relaxation matrix derived distance restraints 131—144
Structure determination, restraints 183
Structure, complex between mRNA and two tRNAs 372—373
Structure, RNA 229—245
Structure, selection 237
Sugar pseudorotation phase angles, in conformations 188
Sugar puckering influence on A to B equilibrium 307 310
Sugar puckering intraresidue spin-spin coupling patterns 97—100
Sugar puckering, A and B forms of DNA 307
Sugar puckering, bent DNA 151
Sugar puckering, Biograf 156
Sugar puckering, equilibrium 110
Sugar puckering, proton-proton scalar coupling constants 181
Sugar puckering, transitions from B to A 265
Sugar puckers, A and B forms of DNA 309
Sugar puckers, energetically unfavorable 191
Sugar puckers, equilibrium distribution 298
Sugar puckers, nucleic acid data 49—52
Sugar puckers, parameters 107
Sugar ring, conformation 4
Sugar, conformational adjustments 115
Supercoiling, circular DNA molecules 11—12
Superhelix, finding the radius 389
Superimposition, nab 384
|
|
 |
| Реклама |
 |
|
|
 |
|
О проекте
|
|
О проекте


 mispair
mispair  pairing
pairing 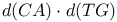
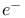 density
density