|
|
 |
| јвторизаци€ |
|
|
 |
| ѕоиск по указател€м |
|
 |
|
 |
|
|
 |
 |
|
 |
|
| Fersht A. Ч Structure and Mechanism in Protein Science |
|
|
 |
| ѕредметный указатель |
Intramolecular catalysis, entropy and 68Ч69
Intramolecular catalysis, versus intermolecular catalysis 69Ч70
Intrinsic binding energy 341
Intron 25 26
Ionization constants of acids and bases 169Ч173
Ionization constants of acids and bases, extraction of 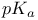 from equations 173 from equations 173
Ionization constants of acids and bases, kinetic 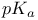 179 179
Ionization constants of acids and bases, macroscopic 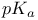 179 179
Ionization constants of acids and bases, microscopic 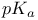 179 179
Ionization constants of groups in enzymes 170 468
Ionization constants of groups in enzymes, determination from rates of chemical modification 187
Ionization constants of groups in enzymes, determination from rates of irreversible inhibition 187 278
Ionization constants of groups in enzymes, determination from steady state kinetics 174Ч176 182Ч184
Ionization constants of groups in enzymes, direct determination 184Ч187
Ionization constants of groups in enzymes, effect of surface charge 179Ч180
Ionization constants of groups in enzymes, effects of environment 187Ч189
Ionization constants of groups in enzymes, highly perturbed 188 189
Isocitrate dehydrogenase 294 459
Isoleucyl-tRNA synthetase 237Ч239 378 379 385 387Ч389
Isoleucyl-tRNA synthetase, molecular mechanism of editing 388 389
Isomorphous replacement 4
Isosteric 378 425 427
Isothermal titration calorimetry (ITC) 207
Isotope exchange kinetics 47 235 236 240 563Ч569
Isotope incorporation 489
Isozyme 461Ч465
Jellyroll 20 21
Jigsaw model 576 583
K system 294
K system, structural explanation 311
Kallikrein 475
kinesin 317 318
Kinetic 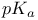 178 178
Kinetic equivalence 94Ч96 482 555
Kinetic isotope effects 96Ч99 235 284 490 498
Kinetic isotope effects, hydride transfer reactions 97 98 463Ч465
Kinetic proofreading 398
Klenow enzyme 405 413
Koshland Ч Nemethy Ч Filmer mechanism 295Ч297
Label-free optical detection 199
Lactate dehydrogenase 465Ч469 472
Lattice simulations 597
Leaving group 81 86 88Ч93
LevinthalТs paradox 575 576 598 599 600
Linear free-energy relationships 58 62Ч65 86 87 599
Linear free-energy relationships, protein engineering of enzymes and 442Ч444
Linear free-energy relationships, protein folding and 579 581 582
Lineweaver Ч Burk plot 111 112 114 120
Linked assays 196
Linked equilibria 127Ч129
Lipoic acid 37
Liver alcohol dehydrogenase see УAlcohol dehydrogenaseФ
Lock-and-key hypothesis 354 369
Loops 20 51
Loops, kinetics of formation 541 569
Lysozyme 23 32 33 43 44 50 497Ч500
Lysozyme, 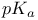 Тs of groups at active site 186 189 Тs of groups at active site 186 189
Lysozyme, affinity labeling 282
Lysozyme, association rate constants 154 165
Lysozyme, crystal and solution structure 50
Lysozyme, electrostatic effects 60 61 499Ч500
Lysozyme, enzyme-substrate complex 44
Lysozyme, evolution 33
Lysozyme, exons 33
Lysozyme, kinetic isotope effects 99
Lysozyme, kinetics of ligand binding to 154 165
Lysozyme, lack of accumulation of intermediates 374
Lysozyme, nonproductive binding 113Ч116 371
Lysozyme, oxocarbenium ion 57 58 60 359 498
Lysozyme, pH dependence of catalysis 497Ч500
Lysozyme, stereochemistry of reaction 255 497
Lysozyme, strain 358 372Ч374 497Ч500
Lysozyme, subsites 44 371
Lysozyme, substrate distortion 358 372Ч374 497Ч500
Lysozyme, tertiary structure 23
Lysozyme, transition stage analogues 358 359
Lysozyme, transition state of reaction 60 61
Lysozyme, van der Waals binding energy of D subsite 329
Macroscopic 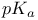 179 179
Malate dehydrogenase 465Ч469
Malate synthase 258
Mass action ratio 308
Mass spectroscopy 242Ч243
Meander 20
Mechanism, definition 216
Mechanism-based inhibitors see УSuicide inhibitorsФ
Metabolite concentrations in vivo 366 367
Metal ion catalysis 74Ч77
Metal ion catalysis, electrophilic 74Ч77 92 392Ч394 462 463 483Ч486
Metal ion catalysis, metal-bound hydroxyl (alkoxide) ion 76 77 392Ч394 483
Metalloenzymes 74Ч77 460Ч463 473 483Ч486
Metaphosphate 259
Methionyl-tRNA synthetase 387
Methyl-methylene transformations 256Ч260
Methylene group (prochiral) 256 257
Michael addition reactions 272 277 284 285
Michael is complex 106
Michaelis constant (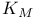 ) 105Ч109 ) 105Ч109
Michaelis constant (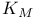 ), evolution of 363Ч368 ), evolution of 363Ч368
Michaelis constant (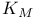 ), meaning of 109 ), meaning of 109
Michaelis constant (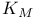 ), pH dependence of 169 174Ч176 ), pH dependence of 169 174Ч176
Michaelis Ч Menten equation 104Ч109
Michaelis Ч Menten equation, affinity labels 277
Michaelis Ч Menten equation, breakdown 119
Michaelis Ч Menten mechanism 105Ч107
Michaelis Ч Menten mechanism, pH dependence 173Ч175
Michaelis Ч Menten mechanism, specificity 380Ч383
Michaelis Ч Menten mechanism, thermodynamic analysis 349Ч355
Microscopic 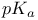 179 179
Microscopic reversibility 93 94 493 500
Minichaperones 606 607 609 610
Modular structure of proteins 33 34
Module 33 577 596 602
Molecular chaperones 603Ч610
Molecular clock 315
Molecular dynamics simulation 537 574 583 584 589 591 592 597
Molecular isotope exchange 266
Molecular tumbling 46
Molten globule state 520 599 600
Monoamine oxidases 284 285
Monod Ч Wyman Ч Changeux mechanism 292Ч296 300Ч304
Monod Ч Wyman Ч Changeux mechanism, versus Koshland Ч Nemethy Ч Filmer mechanism 303Ч304
Monte Carlo methods 597
Motif 9
Motor proteins 317Ч321
mRNA 26 404 413 415
Multi-point attachment theory 248
Multienzyme complexes 34Ч38
Multiproduct analogue 360 361
Multisubstrate analogue 360 361
Multisubstrate systems 119Ч122
Mutagenesis 413Ч416
Mutagenesis, deletion 445
Mutagenesis, oligodeoxynucleotide-directed 413 414
Mutagenesis, PCR 413 414
Mutagenesis, random 415 416 418
Mutagenesis, site-specific 413Ч415
Mutations, choosing 425Ч427
Mutations, isosteric substitution 427
Mutations, mutator strain of T4 phage 390 391
Mutations, nondisruptive deletion 426 560
Myoglobin 11 22 51 289 290 305Ч307
myosin 317 318
NAD-pyruvate 467
NAD-pyruvate, stereochemistry of oxidation-reduction 249 250 548 459
NAD-pyruvate, structure 249
NADH extinction coefficient 192
NAG, NAM 43
Negative cooperativity 296 297
Net rate constants 122Ч125
| Neutron diffraction 6 47
Nicotinamide adenine dinucleotide see У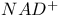 Ф Ф
Nitrocellulose filters 205
Non-Arrhenius kinetics 555 598
Nonbonded interactions 325Ч332
Nondisruptive deletion 426 560
Nonequilibrium dialysis 202
Nonproductive binding 114Ч118 371Ч372
Nonproductive binding, effect on equilibria on enzyme surface 118
Nonproductive binding, effect on ionization constants 177
Nonproductive binding, effect on specificity 372 381
Nonproductive binding, effect on steady state kinetics 114Ч118
Normal distribution 210
Nuclear magnetic resonance (NMR),  relaxation 49 relaxation 49
Nuclear magnetic resonance (NMR), chemical shift 7
Nuclear magnetic resonance (NMR), determination of chirality 251 259 265 266
Nuclear magnetic resonance (NMR), determination of dissociation rate constants 138
Nuclear magnetic resonance (NMR), determination of intracellular pH 186
Nuclear magnetic resonance (NMR), determination of ionization constants 185 186
Nuclear magnetic resonance (NMR), determination of rotational correlation times 46
Nuclear magnetic resonance (NMR), determination of structure 7
Nuclear magnetic resonance (NMR), exchange broadening 138
Nuclear magnetic resonance (NMR), line-width analysis 138 541
Nuclear magnetic resonance (NMR), Nuclear Overhauser Effect (NOE) 7
Nuclear magnetic resonance (NMR), protein mobility and 46Ч49
Nuclear magnetic resonance (NMR), rotation of amino acid side chains and 47 48
Nucleation mechanism 575 576 583 586
Nucleation-collapse see УNucleation-condensation mechanismФ
Nucleation-condensation mechanism 583 585Ч588
Nucleophilic attack on carbonyl group 86Ч90
Nucleophilic attack on phosphoryl and sulfuryl groups 89 90 260Ч266
Nucleophilic attack on saturated carbon 90 91
Nucleophilic catalysis 61 84 85
Nucleophilic catalysis, examples with enzymes 45 85 100 101 311
Nucleophilic catalysis, stereochemistry 254Ч256
Nucleophilic groups on enzymes 76 84 85 274Ч276
Nucleophilicity 85Ч92
Nucleotide binding fold 32 459
Off-lattice simulation 597
Oligomer 24
Operon 35
Ordered mechanism 119 120
Oxocarbenium ion 57 58 60 359 498
P-loop see УWalker consensus sequenceФ
Packing density 24
Papain 482
Parallel reactions 149
Parallel reactions, versus single in folding 579Ч582
ParkinsonТs Disease 285
Partitioning of products 225Ч234
PCR 408Ч410 412Ч416
PCR, error prone 415
Penicillinopepsin 486
Pepsin 486Ч491
Pepsin, affinity labeling of carboxylates 280 487
Pepsin, binding energies of subsites 356 357
Pepsin, kinetic constants for peptide hydrolysis 357
Pepsinogen 490
Pepstatin 489 490
Peptide bond 9 10
Peptides, kinetics of folding 569 570
Peptidyl-proline bond isomerization 541 542 591
Peptidyl-prolyl isomerase 454 603
Peptidyl-prolyl isomerase, protein engineering to cyproase 1 454
pH dependence of enzyme catalysis 173Ч180 460 462 473 474 480 486 490 493 494 498 499
pH dependence of irreversible inhibition and affinity labeling 187 278 279 468 487
pH-jump method 222
Phage display see УBacteriophageФ
Phosphofructokinase 309Ч312
Phosphofructokinase, anomeric specificity 252 253
Phosphoglycerate kinase 23 32 268
Phosphoglycerate kinase, double-mutant cycle  -value analysis 594 -value analysis 594
Phosphoglycerate mutase 119 120
Phosphoramidon 484
Phosphorothioate group 264
Phosphory lase kinase 312Ч314
Phosphory lation/dephosphorylation for control 312Ч315
Phosphoryl transfer reactions 259Ч261
Phosphoryl transfer reactions, stereochemistry 259Ч266 267 268
Phosphorylases 45 312Ч315
Photoaffinity labeling 280
PI3 kinase SH3 domain, folding kinetics 552
Picket fence model for heme 307
Ping-pong kinetics 119Ч121
Plasmid 412
Plasmid, pBR322 412
Plasminogen activator inhibitor 33
Plotting kinetic and binding data 199Ч201 207Ч209
Plotting kinetic and binding data, 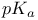 Тs 181 182 Тs 181 182
Plotting kinetic and binding data, computer fitting 209
Plotting kinetic and binding data, first order reactions 199 200
Plotting kinetic and binding data, Michaelis Ч Menten kinetics 201
Plotting kinetic and binding data, second order reactions 200
Poisson distribution 212 410
Polarizability 328 329
Polymerase chain reaction see УPGRФ
Polymerization kinetics 124 390Ч392
Positional isotope exchange 265 266
Pre-steady state 103
Pre-steady state kinetics 132Ч158
Pre-steady state versus steady-state kinetics 216 217
Primary structure 3 4
Principle of maximization of 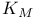 at constant at constant 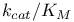 363 364 363 364
Procarboxypeptidase 486
Procarboxypeptidase A2 activation domain,  -value analysis 594 -value analysis 594
Procarboxypeptidase A2 activation domain, folding kinetics 552 594
Processive polymerisation 410
Prochirality 245 247 250 257 262Ч266
Proflavin 220 221
Proline cis-trans equilibrium 9 10
Proline racemase 359
Promoter 412
Proofreading see УEditingФ
Protease A (Streptomyces griseus) 28 475 477
Protease G (Streptomyces albus) 483
Proteases 472Ч491
Protein disulfide isomerase 603
Protein engineering 401 420Ч456
Protein engineering, 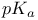 180 327 180 327
Protein engineering,  -value analysis 558Ч563 -value analysis 558Ч563
Protein engineering, Bronsted equation and catalysis 442Ч444
Protein engineering, Bronsted equation and protein folding 579Ч582 590
Protein engineering, changing specificity 452Ч454 465 468 469 481 482
Protein engineering, construction of hetero-oligomers 446Ч449
Protein engineering, determination of energetics of protein structure 523Ч533
Protein engineering, differential and uniform binding energy changes 438 439
Protein engineering, dissection of catalytic triad of serine proteases 450Ч452
Protein engineering, domain structure analysis 445Ч449
Protein engineering, evidence for enzyme-intermediate state complementarity 430Ч432
Protein engineering, evidence for enzyme-transition state complementarity 362 428Ч430
Protein engineering, evolution of activity 438Ч442
Protein engineering, examples 180 297 310 339 480 481 496
Protein engineering, half-of-the-sites activity 448
Protein engineering, hydrogen exchange kinetics 566
Protein engineering, increasing protein stability 532
Protein engineering, kinetic isotope effects 465
Protein engineering, linear free energy relationships with binding energy changes 442Ч444
Protein engineering, protein stability changes 523
Protein engineering, reverse genetics 438Ч442
Protein engineering, surface charge 180 327
Protein kinases 312 314
Protein stability,  helix 523Ч532 helix 523Ч532
Protein stability,  sheet 528 529 532 sheet 528 529 532
Protein stability, engineering increases in 532 535 536
Protein stability, stability and activity 536
Protein, biosynthesis, errors in 385 424
Protein, determination of concentration 214
Protein, diversity 25
Protein, flexibility 32Ч39
Protein-protein recognition 308Ч309
|
|
 |
| –еклама |
 |
|
|
 |
|
ќ проекте
|
|
ќ проекте



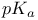 from equations
from equations 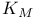 )
) 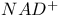 Ф
Ф relaxation
relaxation  -value analysis
-value analysis 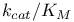
 helix
helix  sheet
sheet