Авторизация
Поиск по указателям
Lesk A.M. — Introduction to Protein Architecture
Обсудите книгу на научном форуме Нашли опечатку?
Название: Introduction to Protein ArchitectureАвтор: Lesk A.M. Аннотация: Written in a clear and engaging style, and profusely illustrated with superb computer graphics, Introduction to Protein Architecture is a textbook for second and third year undergraduate students and beginning post-graduate students, and will be of interest to all biological and medical scientists whose work touches on proteins.
Язык: Рубрика: Биология /Статус предметного указателя: Готов указатель с номерами страниц ed2k: ed2k stats Год издания: 2001Количество страниц: 347Добавлена в каталог: 16.03.2007Операции: Положить на полку |
Скопировать ссылку для форума | Скопировать ID
Предметный указатель
222 62 64 195 201 291 284ff 182 33 60—63 67 326 293 296 70 74 152ff 207ff 213 217 154—157 213—216 154ff 163 70 74 79 115 116 76 240 246 97ff 184 328 143ff 233 9 34 59—61 67—69 71—73 233 77 146 108 62 65—66 17E8, L3 canonical structure (mouse) [1EAP] 243 434 repressor, N-terminal domain (phage 434) [1R69] 309 6-phosphogluconate dehydrogenase (sheep), NAD-binding domain [1PGO] 221 224 Ab1 tyrosine kinase (mouse) / peptide complex [1ABO] 82 Accessible surface area 27 56 Actinidin 183 Actinidin (kiwi fruit) [2ACT] 178 Actinidin (kiwi fruit) [2ACT], Type I’ hairpin loop 97 Acy1 phosphatase (cow) [2acy] 91 Adenylate kinase (pig) [3adk] 332 Alanine racemase (Bacillus stearothermophilus), N-terminal domain [1SFT] 84 Alanine scan 19 Alcohol dehydrogenase (Horse liver), NAD-binding domain [6ADH, 2OHX] 27 218 223 224 Alignment 88 Alignment table, globins 168—169 Alignment table, insulins 171 Allumwandlung (replacement of an amino acid by all 19 others) 19 182 Amino acids 20—22 AN02 (mouse), L3 canonical structure [1BAF] 243 Anti-gp41 monoclonal antibody (human), L3 canonical structure [1DFB] 243 Anti-hypertensive, anti-viral protein (sea anemone) [1BDS] 316 Antibody, B1—B8 245 Antibody, B1—B8, catalytic 19 Antibody, B1—B8, HyHEL-5 238—239 Antibody, B1—B8, McPC603 238—239 Antibody, B1—B8, therapeutic 19 Antigen-binding 236 Antithrombin (human) [1AZX] 294 Apopain (human) [1PAU] 332 Ark clam globin (Scapharca inaequivalvis) [4SDH] 136 Ascorbate oxidase (Escherichia coli) [1ASA] 148 Aspartate carbamoyltransferase (aspartate transcarbamylase) (Escherichia coli) [8ATC] 328 ATPase 3 302 Avian pancreatic (turkey) [1PFI] 322 Azurin 183ff Azurin (Alcaligenes denitrificans) [2AZA] 179 B-factor 38 B1—B8, antibody 245 B35, class I MHC protein (human) [1A1N] 251 254 B53, class I MHC protein (human) [1A1O] 255 257 258 Bacteriochlorophyll 10 Bacteriochlorophyll a protein (Prosthecochloris aestuarii) [4BCL] 327 Bacteriorhodopsin (Halobacterium halobium) [1BRD] 139 Barley chymotrypsin inhibitor [2CI2] 83 Barnase (Bacillus amyloliquefaciens) [1BRN] 87 Barrel see “ Barstar (Bacillus amyloliquefaciens), 68 71 Binding change mechanism of ATPase 302—303 Bioinformatics 16 Bovine liver phosphotyrosine protein phosphatase [1PHR| 190—191 Bovine pancreatic trypsin inhibitor (and trypsin) [1TPA] 108 Bovine pancreatic trypsin inhibitor [5PTI] 82 C-Ha-ras encoded p21 (human), catalytic domain [4Q21] and [6Q21] 87 Caenorhabditis elegans genome 16 94 Canonical structures, immunoglobulin 240ff Canonical structures, T-cell receptor 262 Carboxypeptidase a (cow) [5CPA] 327 Carboxypeptidase a (cow) [5CPA], helix 142 Carboxypeptidase a (cow) [5CPA], helix-helix contact 163 164 Carboxypeptidase a (cow) [5CPA], IIbcellobiose (Escherichia coli), A chain [1IIB] 190—191 CASP (Critical Assessment of Structure Prediction) 90 Catalytic antibody 19 Catalytic triad (Ser-His-Asp) 204 207 CATH (class, architecture, topology, homologous superfamily) 43 127 129ff CDR see “Complementarity-determining region” Chaperone 3 297 Chemical shift 41 Chymotrypsin (cow) [8GCH] 204 Chymotrypsin (cow) [8GCH], specificity pocket 213 Chymotrypsin inhibitor 2 (barley) [2CI2] 323 Citrate synthase 281ff Citrate synthase (chicken) [3CTS, 5CTS] 138 283 Cleaved 292 296 Complementarity-determining region (CDR) 233 236 240ff Complementarity-determining region (CDR), T-cell receptor 262 Concanavalin A (jackbean) [3CNA] 147 Conformation 31-32 Conformational angles 31—32 35 Core of protein structure 18 50 88 180—181 Cow mitochondrial F1-ATPase [1BMF] 304 Crambin (Crambe abyssinica) [1CRN] 24 Critical Assessment of Structure Prediction (CASP) 90 Cryo-EM 42 Crystal-packing forces, effect on structure 180 Cytochrome 324 Cytochrome 310 Cytochrome 103 Cytochrome c (rice) [1CCR] 26 138 Cytochrome c (tuna) [5CYT] 138 Cytokine: glycosylation-inhibiting factor (human) [1GIF] 329 D-ribose-binding protein (Escherichia coli) [2DRI] 333 D1.3 (mouse) binding hen egg white lysozyme [1VFB] 86 DALI 43 88 127 131 Dali Domain Dictionary 131 Databanks 45—46 Death domain of p75 low affinity neurotrophin receptor (rat) [1NGR] 312 Denaturation 37 Dihydropteridine reductase (rat), NAD-binding domain [1DHR] 221 223 Direct methods of phase determination 38 Discs large protein, PDZ3 domain, DHR3 domain (human) [1PDR] 318 Disulphide bond 23 DNA 61 DNA complex of transcription factor Skn-1 (Caenorhabditis elegans) [1SKN] 120 DNA polymerase 113 Domain 75 Domain swapping 175-176 Drosophila melanogaster genome 16 94 204 Drug design 19 Elastase (pig) [3EST] 215 Elastase (pig) [3EST], type II’ hairpin loop 98 Engrailed homeodomain (Drosophila melanogaster) [1ENH] 80 Enolase 173—174 Erythrocruorin (Chironomus thummi), B/G helix contact [1ECD] 198 Erythrocruorin (Chironomus thummi), B/G helix contact [1ECD], loop 104—105 Exon, proposed relation to protein structural unit 166 F1-ATPase see “ATPase” Fab J539 (mouse), H1 loop [2FBJ] 241 Fab J539 (mouse), H1 loop [2FBJ], H3 loop 246 Fab KOL (human), CH1 domain [2FB4] 234 Fab KOL (human), VL domain 234 Fab McPC603 (mouse), H3 loop [1MCP] 246 Fab McPC603 (mouse), H3 loop [1MCP], antigen-binding site 238 Fab McPC603 (mouse), H3 loop [1MCP], VH domain 244 Fab TE33 (mouse), antigen-binding site [1TET] 245 Farnesyl diphosphate synthase (chicken) [1FPS] 315 Fatty acid binding protein (human) [1HMS] 320 Ferredoxin (Azotobacter vinelandii) [6FD1] 159 Ferredoxin reductase (spinach) [1FNB] 134 Fibre diffraction 36 FK506-binding protein (human) [2FKE] 86 FK506-binding protein (yeast) [1YAT] 86 Flavodoxin (Clostridium beijerinckii) [5NLL] 29 128 133 Fold recognition 90 Folding pathway 23 Formate dehydrogenase (Pseudomonas sp. 101) [2NAD] 219 Four-helix bundle 132 135ff Fructose permease subunit IIb (Escherichia coli) [1BLE] 331 FSSP (Fold classification based on Structure-Structure alignment of Proteins) 88 Fv D1.3 (mouse) [1VFB] 86 GCN4-bZIP, DNA complex (yeast) [2DGC] 139 Gene duplication 173 genetic code 17 Genomics 12 15—16 94 Globin (Aplysia limacina) [1MBA] 202 Globin (Glycera dibranchiata) [1HBG] 136 Glucoamylase fragment (Aspergillus awamori) [1GAI] 316 Glutathione peroxidase (cow) [1GP1] 333 Glyceraldehyde-3-phosphate dehydrogenase (Bacillus stearothermophilus), NAD-binding domain [1GD1] 220 Glycolate oxidase (spinach) [1GOX] 75 153 156—157 Glycosyl-asparaginase domain (Flavobacterium meningosepticum) [1PGS] 145 Green fluorescent protein (Aequorea victoria) [1EMA] 321 GroEL (Escherichia coli) [1DER] 298 GroEL-GroES 3 297 GroEL-GroES complex (Escherichia coli) [1AON] 300 301 Haemoglobin (horse), 198 Haemoglobin (human) [1HHO] 285 288 289 Haemoglobin (human) [1HHO], a-chain, haem pocket 200 Haemoglobin (human) [4HHB] 137 Haemoglobin S 182 Haemoglobin, 284ff Haemoglobin, alignment table 168—169 Haemoglobin, allosteric change 284ff Haemoglobin, evolution 166ff 195ff Haemoglobin, oxygen binding 285 Hairpin see “ Haplotype 249 Haptoglobin 174 Helical wheel 67 Helices 9 59 Helix cap 66 Helix capping 62 66 Helix hairpin 78 Helix interface see “Helix-helix packing” Helix, 62 64 195 Helix, 33 60—63 67 Helix, 62 65—66 Helix, conformational parameters 67 Helix, polyproline II 62 67 68 Helix-helix packing 141ff 163—164 201 203 Helix-interface-shear mechanism of conformational change 277ff Hinge motion 274 HLA-B53, MHC protein (human) [1A1M] and [1A1O] 250 Homology modelling 18 90 184ff Human genome 16 94 Hydrogen bond pattern 59 Hydrogen bond pattern, effect of proline 62 Hydrogen bond pattern, helix 59ff Hydrogen bond pattern, NAD-binding domains 218ff Hydrogen bond pattern, serine proteinases 210 215 Hydrogen bond pattern, sheet 68ff Hydrogen bonding 23 27 Hydrophobic effect 21 49 62 Hydrophobicity scale 21 22 HyHEL-5 (mouse), hen egg white lysozyme complex [1BQL] 239 HyHEL-5 (mouse), hen egg white lysozyme complex [1BQL], L3 canonical structure [3HFL] 242 HyHEL-5, antibody 238—239 I-Ak, class II MHC protein (human) [1IAK] 252-254 260 Immunoglobulin D1.3 Fv fragment (mouse) [1FVB] 86 Immunoglobulin G MAB231 (mouse) [1IGT] 232 Immunoglobulin HyHEL-5 Fab fragment (mouse), hen egg white lysozyme complex [1BQL] 239 Immunoglobulin HyHEL-5 Fab fragment (mouse), hen egg white lysozyme complex [1BQL], L3 canonical structure [3HFL] 242 Immunoglobulin J539, Fab fragment (mouse) [2FBJ] 86 Immunoglobulin McPC603, Fab fragment (mouse) [1MCP] 102 Immunoglobulin McPC603, Fab fragment (mouse) [1MCP], hairpin loop 101 104 Immunoglobulin structure 231ff Immunoglobulin superfamily 230 Immunoglobulin TE33, Fab fragment (mouse) [1TET] 86 245 Immunoglobulin TE33, Fab fragment (mouse) [1TET], CL domain 69 72 Immunoglobulin, antigen-binding by 236 245 Immunoglobulin, B1-B8 244 Immunoglobulin, canonical structures 240ff Immunoglobulin, gene assembly 229 244 Immunoglobulin, HyHEL-5 238—239 Immunoglobulin, McPC603 238—239 Immunoglobulin, TE33 245 Induced fit 258 Influenza haemagglutinin [3HMG] 149 Insulin (pig) [1ZNI] 24 Insulin (pig), 2-zinc form, dimer [3INS] 278 282 Insulin (pig), 2-zinc form, hexamer 279 Insulin (pig), 2-zinc form, monomer 280 Insulin (pig), 4-zinc form, dimer [1ZRO] 24 278 Insulin (pig), 4-zinc form, hexamer 279 Insulin (pig), 4-zinc form, monomer 280 Insulin, alignment table 171 Insulin, conformational change 277 Insulin, zinc binding sites 105ff Interleukin-5 (human) [1HUL] 176 Internet 16 Irregular structures 158 Isocitrate dehydrogenase (Escherichia coli) [5ICD] 54 J539 (mouse) [2FBJ] 86 J539 (mouse) [2FBJ], H3 loop 246 J539 (mouse) [2FBJ], L3 canonical structure 242 Keratin 9 15 Lactate dehydrogenase 174 Lactate dehydrogenase (pig), NAD-binding domain [9LDT] 223 Lactoferrin (human) [1LFG] and [1LFH] 274 Lactoferrin, hinge motion 274 Leghaemoglobin (lupin) [2LH7] 177 196 Leghaemoglobin (lupin) [2LH7], F/H helix contact 198 Leghaemoglobin (lupin) [2LH7], haem pocket 200 Lens crystallins 173 Light-harvesting protein: peridinin-chlorophyll protein (Amphidimum carterae) [1PPR] 314 Lithostatine: pancreatic stone inhibitor (human) [1LIT] 323 Lock and key model of enzyme specificity 36 Loops 95ff Loops, classification 96 Loops, sequence-structure relationships 97ff 184 Low temperature electron microscopy 42 Lysozyme (hen egg white) [1HEW] 106 Lysozyme (hen egg white), HyHEL-5 (human) complex [1BQL] 239 Lysozyme (phage 81 130 326 Lysozyme (phage 183 MAD (multiwavelength anomalous dispersion) 38 Major histocompatibility complex (MHC) proteins 247 263ff Major histocompatibility complex (MHC) proteins, binding of peptides 25lff 255ff Major histocompatibility complex (MHC) proteins, interaction with TCR 263ff Malate dehydrogenase 174 Malate dehydrogenase (Escherichia coli), NAD-binding domain [1EMD] 220 223 Malate dehydrogenase (pig), NAD-binding domain [1MLD] 84 Mannose-specific agglutinin (snowdrop) [1JPC] 319 Max protein, DNA-binding domain (mouse) [1AN2] 313 McPC603 (mouse), H3 loop [1MCP] 246 McPC603 (mouse), H3 loop [1MCP], antigen-binding site [2MCP] 238 McPC603 (mouse), H3 loop [1MCP], hairpin loop 101ff McPC603 (mouse), H3 loop [1MCP], VH domain 244 McPC603, antibody 238—239 MHC see “Major histocompatibility complex” MHC protein/T-cell receptor complex (mouse) [2CKB] 263—265 Modular protein 75 Molecular clock 170 Molecular replacement 38 Monellin (african serendipity berry) [4MON] 325 Motif 95 218 Motif, canonical structures of antigen-binding loop 243 Multiwavelength anomalous dispersion (MAD) 38 Murine/human ubiquitin-conjugating enzyme ubc9 [1U9B] 150
Реклама
 |
|
О проекте
|
|
О проекте



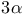 ,
, 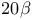 -hydroxysteroid dehydrogenase, NAD-binding domain (Streptomyces hydrogenans) [2HSD]
-hydroxysteroid dehydrogenase, NAD-binding domain (Streptomyces hydrogenans) [2HSD] 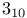 helix
helix  transition (serpins)
transition (serpins) 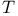 transition (haemoglobin)
transition (haemoglobin) 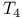 lysozyme, effect of mutation
lysozyme, effect of mutation  -Helix
-Helix 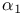 -antitrypsin (human) [2PSI]
-antitrypsin (human) [2PSI]  -barrel
-barrel  unit
unit  -crystallin domain (cow) [1AMM]
-crystallin domain (cow) [1AMM] 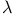 cro-DNA complex
cro-DNA complex 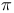 -helix
-helix  ,
,  [1BRS]
[1BRS]  (cow) [3B5C]
(cow) [3B5C]  (Desulfovibrio vulgaris Miyazaki) [2CDV]
(Desulfovibrio vulgaris Miyazaki) [2CDV]