јвторизаци€
ѕоиск по указател€м
Helmreich E.J.M. Ч Biochemistry of Cell Signalling
ќбсудите книгу на научном форуме Ќашли опечатку?
Ќазвание: Biochemistry of Cell Signallingјвтор: Helmreich E.J.M. јннотаци€: The Biochemistry of Cell Signalling deals in depth with the principles of cell signalling, concentrating on structure and mechanism. It will serve as a reliable map through the maze of cell signalling pathways and help the reader understand how malfunctions in these pathways can lead to disease. The book is divided into four parts. Part 1 describes the machinery of signal transduction starting with the properties of signals, receptors (including receptor activation), regulators, and the molecules that link receptor and regulator. The design of signalling cascades is explained by describing central signalling pathways: the Ras-regulated MAPK and PI-3 pathways; the Rho/Rac/Cdc 42 pathway controlling chemotaxis and regulating the cytoskeleton; the G protein coupled receptor cascades in response to sensory and hormonal signals; signalling by TGF-ss in morphogenesis; cytokine signalling that controls haemopoiesis. There is also a discussion of the insulin response. As phosphorylation - dephosphorylation is involved in nearly all cellular regulatory processes, Part 1 concludes with a synopsis of its role in signalling. Part 2 describes the implementation of the signalling cascades focusing on the effect on gene transcription. After a brief description of the transcriptional machinery the regulation of transcription by cytokines and growth factors in the control of cell growth and the mechanisms and sites of control are discussed in detail. The regulators discussed include Jun/Fos, NF-AT, SREBPs, and STATs. The next two chapters cover gene regulation by nuclear receptors, including both the steroid hormone receptors and non-steroid nuclear receptors e.g. the retinoic acid receptors RAR and RXR. Part 3 studies the global cellular regulatory programs for the control of cell growth and proliferation. The first chapter concerns the regulation of the cell cycle and the role of the cyclin-dependent kinases, telomerase, Ran, and cell cycle checkpoints. The next topic is the signalling pathways in apoptosis: the TNF-receptor family death receptors, caspases, and the intracellular apoptosis signals and the role of apoptosis in the lifecycle of cells. Part 3 ends with a discussion of the signal pathways involved in the immune response, focusing on the involvement of cell-cell interactions. Part 4 considers loss of regulatory control and its consequences with respect to the molecular basis of cancer. It first describes the cellular regulatory proteins that have oncogenic potential, how they can become oncogenic and cause the transformation of normal cells to cancerous cells. Next is an analysis of the loss of developmental controls, the APC protein, ss-catenin, and the Wnt pathway, that lead to mature terminally differentiated cells reverting to immature embryonic cells. The book ends with a summary of the molecular and cellular causes of cancer and an outlook for novel therapies. Throughout the text, the emphasis is on structure and mechanism and is well illustrated with 200 figures. The Biochemistry of Cell Signalling will be an invaluable companion to all graduate students studying cell signalling.
язык: –убрика: Ѕиологи€ /—татус предметного указател€: √отов указатель с номерами страниц ed2k: ed2k stats √од издани€: 2001 оличество страниц: 357ƒобавлена в каталог: 22.12.2005ќперации: ѕоложить на полку |
—копировать ссылку дл€ форума | —копировать ID
ѕредметный указатель
287Ч292 77-78 81Ч82 78 5-HT (serotonin) receptor 77 Abl tyrosine kinases 38 Activins 102 103t ADAM proteins 5Ч6 Adaptor proteins and linkers, ankyrin repeats 37 38 Adaptor proteins and linkers, G proteins 43Ч45 Adaptor proteins and linkers, notch repeats 37 Adaptor proteins and linkers, PH domain plate 4 (2p) 35Ч36 Adaptor proteins and linkers, phosphotyrosine phosphatases 37Ч38 41Ч43 Adaptor proteins and linkers, PTB domain 35Ч36 37 Adaptor proteins and linkers, Ras 46Ч49 50 Adaptor proteins and linkers, SH2 domain plate 2 (lp) 33Ч34 Adaptor proteins and linkers, SH3 domain plate 3 (2p) 34Ч35 Adaptor proteins and linkers, sterile 37 38 Adaptor proteins and linkers, tyrosine kinases plate 5 (3p) 37Ч40 Adenomatous polyposis coli (APC) gene/protein 286Ч292 Adenylate cyclase, adenylyl cyclaseЧ 10 (6p) Aldosterone, structure 195 Allosteric activation, of proteins 132Ч133 Ankyrin repeats 37 38 Antibodies, structure 255Ч256 Antigen presentation plate 28 (15p) 251Ч252 253 254 Antigen receptors plate 29 (16p) 255Ч257 Apoptosis and cancer 243Ч245 Apoptosis and immune system function 245 246 Apoptosis and nervous system development 245 Apoptosis and self-tolerance 263 Apoptosis, prevention plate 27 (15p) 241Ч243 Apoptosis, promotors 234Ч241 Arrestin and G-protein coupled receptor signalling 83 Arrestin and receptor desensitisation 81Ч82 83 B cells, activation 250Ч251 B cells, antigen receptors 255 B cells, signalling pathways 258Ч259 B cells, survival and death 261Ч263 Basic helix-loop-helix (bHLH), motif 162Ч163 Basic helix-loop-helix (bHLH), structure plate 17 (10p) Bax and apoptosis 241 Bax and cancer 243Ч245 Bcl-2 and apoptosis 241Ч243 Bcl-2 and cancer 243Ч245 Bcl-2- 27 (15p) Bone morphogenetic proteins (BMPs) 102 103 103t 104 Brain-derived neurotrophic factor (BDNF) 14 15t Breast cancer, and oestrogen receptor 197 199 Calcineurin, structure plate 13 (7p) 126 cAMP and G-protein coupled, receptors 78 79 cAMP and G-protein coupled,i n olfaction 95Ч96 cAMP response-element-binding protein (CREB) 174 Cancer, and adenomatous polyposis coli (APC) gene 286Ч287 Cancer, and adenomatous polyposis coli (APC) protein 287Ч292 Cancer, and apoptosis 243Ч245 Cancer, and DNA viruses 299Ч300 Cancer, and loss of developmental control 284Ч285 Cancer, and retroviral infection 270Ч271 299 Cancer, inherited cancer genes 294Ч295 Cancer, oncogenes 269Ч276 Cancer, proto-oncogene mutations 295Ч299 Cancer, tumour-suppressor genes 276Ч282 Caspases plate 26 (14p) 237Ч238 239 Caveolae, and glucose transport 141 143 CBL protein, and cancer 273 CD (cluster of differentiation) receptors, and immune response 256Ч257 CD95 receptor, and apoptosis 235 Cdc42 GTPase 64Ч65 Cdc42 GTPase, and cytoskeleton assembly 65Ч70 Cdc42 GTPase, and Wiskott Ч Aldrich syndrome protein (WASP) 73 Cdc42 GTPase, control of phospholipases 70Ч72 Cell adhesion, and cancer 287Ч289 Cell adhesion, role of integrins 68t 69Ч70 Cell cycle, checkpoint control 228Ч230 295Ч299 Cell cycle, chromosome duplication 225 226 Cell cycle, cyclin-dependent kinases see УCyclin-dependent kinases (Cdks)Ф Cell cycle, cyclins 214 215 Cell cycle, cytokinesis 226Ч228 Cell cycle, DNA replication 224Ч225 Cell cycle, evolution of 230 Cell cycle, phases 213 Cell death, programmed see УApoptosisФ Cell lineage differentiation, cytokine functions 117Ч119 cGMP, in visual response 90Ч92 Chaperones, and steroid hormone receptors 197 198 Chaperonins, and steroid hormone receptors 197 198 Chemotaxis 70Ч72 Chemotaxis, G protein control of 89Ч90 Chromosome duplication 225 226 Colony-stimulating factors (CSFs) 117 118t Colony-stimulating factors (CSFs), processing 7 Cortisol, structure 195 Covalent activation, of proteins 132Ч133 CREB (cAMP response-element-binding protein) 174 Crk adaptor protein 31Ч32 Csk kinase 40 Cyclin-dependent kinases (Cdks) 214 Cyclin-dependent kinases (Cdks), and cell-cycle transitions 216Ч224 Cyclin-dependent kinases (Cdks), and DNA replication 224Ч225 Cyclin-dependent kinases (Cdks), and evolution of the cell cycle 230 Cyclin-dependent kinases (Cdks), cyclin-dependent-kinase inhibitor (CKI) plate 25 (14p) Cyclins 214 215 see Cytochrome c, and apoptosis 238 240 Cytokines, and cell lineage differentiation 117Ч119 Cytokines, and embryonic development 116Ч117 Cytokines, and T cell differentiation 252Ч254 255t Cytokines, antiviral effects 118 119 Cytokines, JAK/STAT signalling pathway 111Ч116 Cytokines, receptors 110Ч111 114 Cytokinesis 226Ч228 Cytoskeleton assembly, role of GTPases 65Ч70 Cytosolic kinases 123 DAF-2, receptor 17Ч18 DAF-2, signalling pathway 148Ч149 Death receptors 235Ч236 237 Development, and steroid hormone receptors 193Ч195 Development, embryonic, cytokine functions 116Ч117 Development, embryonic, STATs 116Ч117 Development, loss of control of, and cancer 284Ч285 Development, of nervous system, and apoptosis 245 Diabetes 137Ч138 DNA tumour viruses 299Ч300 DNA, damage 240Ч241 DNA, histone modification 166Ч167 DNA, methylation 167Ч168 297 DNA, nucleosomes plate 22 (12p) 165Ч166 DNA, replication 224Ч225 DNA, synthesis 62Ч63 DNA-binding proteins, structural motifs plate 15 (9p) plate plate plate plate plate plate 161-4 Ecdysone receptor 192Ч193 Ecdysteroids 192 EGF see УEpidermal growth factor (EGF)Ф Embryonic development, cytokine functions 116Ч117 Eph-like receptors 9t 18 37 38 Ephrins 9t Epidermal growth factor (EGF), EGF receptor and cancer 272 Epidermal growth factor (EGF), signal transduction mechanisms 9t 10Ч11 Erythropoietin (EPO) receptor, ligand-dependent dimerization 25Ч27 Estrogen receptor see УOestrogen receptorФ Fibroblast growth factors (FGFs), signal transduction mechanisms 9t 12Ч14 Focal adhesion kinase (FAK), integrin-FAK signalling system 68Ч70 Fos/Jun transcription factors 172Ч174 G proteins, 8 (5p) G proteins, 7 (4p) G proteins, 79Ч80 G proteins, and cancer 273 G proteins, G- 6 (4p) G proteins, GDP/GTP cycle 43Ч45 G proteins, heterotrimeric G proteins 88Ч90 G proteins, in visual response plate 12 (7p) G proteins, monomeric G proteins 89Ч90 G proteins, Ras/GAP complex structure plate 9 (5p) G proteins, Ras/MAP kinase pathway 57Ч64 G proteins, Rho/Rac/Cdc42 GTPases 65Ч73 G-protein-coupled receptors, and olfaction 92Ч96 G-protein-coupled receptors, and Ras/MAP kinase pathway 83Ч85 G-protein-coupled receptors, and taste transduction 96Ч97 G-protein-coupled receptors, and visual response 90Ч92 G-protein-coupled receptors, control of hormonal signalling 79Ч81 G-protein-coupled receptors, receptor desensitisation 81Ч2 83 G-protein-coupled receptors, rhodopsin activation 85Ч88 G-protein-coupled receptors, second messenger signalling plate 10 (6p) plate 78 79 G-protein-coupled receptors, structure 76Ч78 GATA transcription factors 175 GDNF (glial-cell-line-derived neurotrophic factor) 14Ч17 GDP/GTP cycle, 8 (5p) GDP/GTP cycle, 7 (4p) GDP/GTP cycle, and cell signalling 43Ч45 GDP/GTP cycle, G- 6 (4p) GDP/GTP cycle, Ras/GAP complex plate 9 (5p) Gene transcription see УTranscriptionФ Genomic imprinting 167Ч168 Glial-cell-line-derived neurotrophic factor (GDNF) 14Ч17 Glucose, homeostasis, insulin actions 140Ч142 Glucose, protein glycation 138 139 Glutamic-acid-rich proteins (GARPs), in visual response 91 Glycation, of proteins 138 139 Glycogen phosphorylase, regulation by phosphorylation dephosphorylation 130Ч132 Glycogen phosphorylase, structure plate 14 (8p) Grb2 (growth-factor-receptor-binding protein), SH2 and SH3 domains 32 34Ч35 Growth factor receptors and cancer 272 Growth factors, and cancer 271Ч272 Growth factors, processing 5Ч8 Growth hormone (GH) receptor, binding domain structure plate l (lp) Growth hormone (GH) receptor, ligand-dependent dimerization 24Ч25 GTPase-activating proteins (GAPs), and control of Ras activity 46Ч48 GTPases 43Ч45 GTPases, and cytoskeleton assembly 65Ч70 GTPases, control of phospholipases 69 70Ч72 GTPases, Ras superfamily 64Ч65 GTPases, signalling pathways 72Ч73 Guanine nucleotide release proteins (GNRPs), and control of Ras activity 48Ч49 50 Gusducin, and taste transduction 96Ч97 Haematopoietic cells, differentiation, cytokine functions 117Ч119 Heat-shock protein (Hsp) system, and steroid hormone receptors 197 198 Hepatocyte growth factor (HGF) 9t 18 HGF-like factor 9t Histones 165-6 Histones, modification 166Ч167 Histones, nucleosome core particle structure plate 22 (12p) Homeodomain proteins 162 163 Homeodomain proteins, MAT 15 (9p) Homeotic genes 162 193Ч5 Immune response 250Ч251 259Ч260 Immune response, antigen presentation plate 28 (15p) 251Ч252 Immune response, antigen receptors plate 29 (16p) 255Ч257 Immune response, lymphocyte survival and death 261Ч263 Immune response, lymphoid organs 250 251 Immune response, signalling pathways in T cells and B cells 258Ч259 Immune response, T cell differentiation 252Ч254 255t Immune response, T cell selection 252 Immune surveillance, and apoptosis 246 Immunoglobulins, structure 255Ч256 Imprinting, of genes 167Ч168 Insulin receptor, homologues 17Ч18 Insulin receptor, insulin receptor substrates (IRS) 143Ч146 Insulin receptor, signal transduction mechanisms 9t 17 Insulin receptor, structure 11 Insulin, and glucose homeostasis 140Ч142 Insulin, as growth factor 137 142Ч150 Insulin, functions 137Ч40 Insulin, history 137 Insulin, processing 8 Insulin, signal transduction mechanisms 9t Insulin, signalling pathways 142Ч150 Insulin-like factor, signal transduction mechanisms 9t Integrin-FAK signalling system 68Ч70 Integrins 65Ч70 Interferons 118 119 Interleukin-1 26 (14p) 237 Interleukins, and T cell differentiation 252Ч254 255t JAK/STAT signalling pathway, and cytokines 111Ч116 JNKs (Jun N-terminal kinases) 60 Jun/Fos transcription factors 172Ч174 Juvenile hormone (JH) 195 Keratinocyte growth factors (KGFs) 9t 12 KIT receptor 9t 14 KL ligand 9t 14 Leptin, cross-talk with insulin signalling pathways 148Ч150 Leucine zipper motif 162 163 Ligands, growth factor processing 5Ч8 Linker proteins see УAdaptor proteinsФ Lymphocytes see УB cellsФ УT Macrophage colony-stimulating factor-1 (MCSF-1) 9t Macrophage colony-stimulating factor-1 (MCSF-1), signal transduction mechanisms 11 MAD proteins 176 Major histocompatibility complex (MHC), and antigen presentation 251Ч252 253 254 Major histocompatibility complex (MHC), and antigen receptor, structure plate 29 (16p) MAP kinase pathway see УRas/MAP kinase pathwayФ MAP kinases 60Ч62 MAP kinases, role in growth and proliferation 62Ч63 MAPKKs (MEKs) 60 Mast cell growth factor 9t 14 MAT 15 (9p) MCSF-1 (macrophage colony-stimulating factor-1) 9t Metalloproteinases, and growth factor processing 5Ч6 Methylation, of genes 167Ч168 Methylation, of genes, and cancer 297 MHC see УMajor histocompatibility complex (MHC)Ф Mitochondria, and apoptosis 238 240 Mitogen-activated protein kinases see УMAP kinasesФ Morphogenesis, TGF- 108Ч109 Multi-enzyme organelles, phosphorylation cascades 129Ч130 Nerve growth factor (NGF), and apoptosis 245 Nerve growth factor (NGF), p75NGFR 15Ч16 245 Nerve growth factor (NGF), signal transduction mechanisms 14Ч17 Nervous system development, and apoptosis 245 Neuregulin 9t Neurofibromin 1 Neurofibromin, and control of Ras activity 47Ч48 Neurotrophins 14Ч17 Neurotrophins and apoptosis 245 NF- 164 175 NF- 23 (13p) NF- 18 (10p) NF-AT transcription factor 174Ч175 NF-AT transcription factor, NF-AT/Jun/Fos/DNA complex plate 23 (13p) NF-xB transcription factor 175 NGF see УNerve growth factor(NGF)Ф Notch repeats 37 Nuclear receptors, for non-steroids see УRetinoic acid receptors (RARs)Ф УRetinoid УThyroid Nuclear receptors, for steroid hormones see УSteroid hormone receptorsФ Nuclear transport, and transcriptional control 181Ч182 183 Nucleosomes 165Ч166 Nucleosomes, core particle structure plate 22 (12p) Oestradiol, structure 199 Oestrogen receptor, and breast cancer 197 199 Olfaction, neuronal connections 92Ч93 Olfaction, odorant receptors 93Ч95 Olfaction, odorants 93 Olfaction, signal transduction in olfactory neurons 95Ч96 Oncogenes, and proto-oncogene functions 269Ч271 Oncogenes, and signalling pathways 271Ч273 Oncogenes, transformation from protooncogenes 273Ч276 Oxidative stress, stress activated transcription factors 179Ч180 p53, and cancer 243Ч245 279 280Ч282 p53, structure plate 30 (16p) p75NGFR, and apoptosis 245 p75NGFR, signal transduction mechanisms 15Ч16
–еклама
 |
|
ќ проекте
|
|
ќ проекте



 -catenin, and cancer
-catenin, and cancer 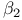 adrenergic receptor
adrenergic receptor  -motif (SAM)
-motif (SAM) 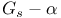 complex plate
complex plate 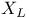 , structure plate
, structure plate 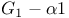 subunit plate
subunit plate 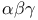 -holocomplex structure plate
-holocomplex structure plate 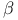 -subunit functions
-subunit functions 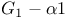 subunit plate
subunit plate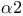 /MCM1/DNA complex structure plate
/MCM1/DNA complex structure plate  B transcription factor
B transcription factor